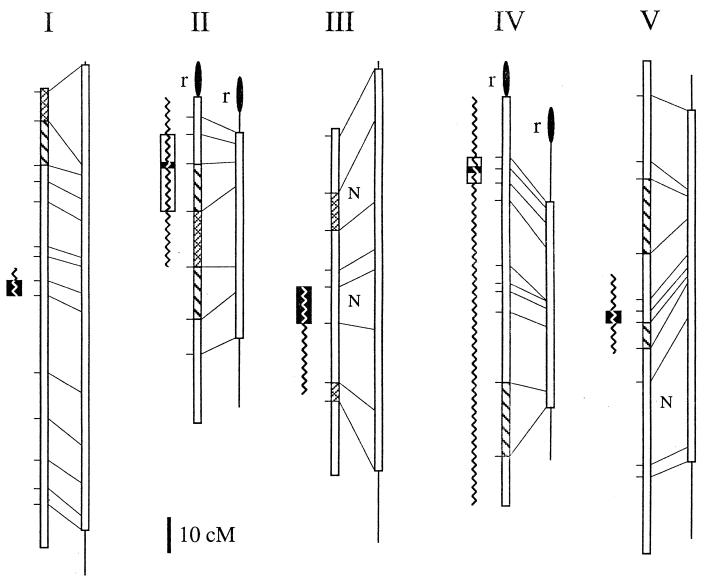
Figure 3.
Genetic maps of Arabidopsis chromosomes I–V. For each column, on the left is the published map from male and female meioses (ref. 12; http://nasc.nott.ac.uk/new_ri_map.html) and on the right is a male-specific meiotic map calculated by using the data presented here (the lines at the extremities of the male-specific map represent unsampled regions). Marker positions (horizontal lines), corresponding markers on both maps (diagonal lines), intervals with observed nonparental ditype tetrads (N), regions of enhanced (cross-hatched boxes) and reduced (hatched boxes) recombination in males, and rDNA (filled ovals) are indicated. Centromere positions as defined by recombinant individuals at the markers assayed (open boxes) and calculated by a mapping function (solid boxes; see Materials and Methods) are shown at the left of each chromosome map. Centromeres for chromosomes I–V were mapped between 7G6 and T27K12 (52–59 cM), m246 and THY1 (11–33 cM), GL1 and NIT1 (45–55 cM), GA1 and nga8 (17–29 cM), and nga76 and PHYC (71–74 cM), respectively. At each centromeric interval, there are 6, 14, 15, 7, and 2 (chromosomes I–V, respectively) recombinants remaining. On chromosomes 2 and 4, no recombinants were detected at mi310 and nga12, respectively. Previous centromere localizations derived from chromosome fragmentation experiments are displayed as jagged lines (35, 36).