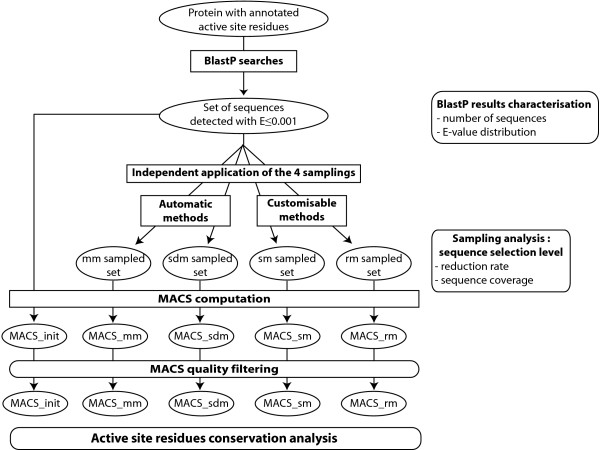
Figure 1.
Strategy flowchart. For each protein in the initial 284 protein dataset, the set of potential homologous sequences was detected by BlastP searches. BlastP results were then characterised according to the number of sequences detected and their associated E-value distribution. The 4 sampling methods (2 automatic methods: the mean method mm and the second derivative method sdm; 2 customisable methods: the strips method sm and the random method rm) were independently applied to the initial set and analysed in terms of reduction rate properties and sequence coverage between the methods. Finally, the 5 associated multiple alignments of complete sequences (MACS) were computed. Taking into account the common high quality MACS, the variation of the information content of the sampled sets were studied, based on the conservation of the active site residues.