FIG. 3.
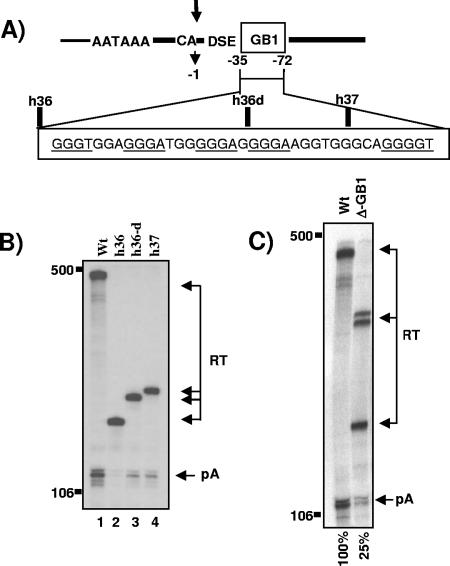
G-rich sequence adjacent to the core poly(A) site is critical for basal cleavage activity. A) The structure of the MC1R poly(A) site with the adjacent G-rich sequence is shown. CA and the arrow indicate the primary site of cleavage, DSE represents the downstream sequence element, and the G-rich sequence is shown as an open box named GB1. The position of the G-rich sequence is indicated with numbers, where the site of cleavage is set to 0 and nucleotides in the 3′ flank are numbered in negative values in a 5′ to 3′ direction. GB1 is enlarged below, the hnRNPH binding protein motifs are underlined, and the deletion clones are indicated above the enlarged sequence. B) RNase protection analysis of total RNA isolated from HEK293 cells transiently transfected with the wt, h36, h36-d, and h37 mutant MC1R reporter plasmids (lanes 1 to 4). pA and RT are the same as described in the legend to Fig. 2B. Note that the length of the protected readthrough bands (RT) differs in each lane, because the introduced deletions overlap with the protection probe, which contains wild-type 3′ flanking sequences. C) Representative RNase protection analysis of transfections with wt and the Δ-GB1 deletion construct. Phosphorimager quantitations of cleavage efficiency calculated as the ratio between cleaved and uncleaved transcripts (pA/RT) are indicated at the bottom of the gel, and the numbers are the mean values of three independent experiments with a standard deviation of ±8.6%. pA and RT are the same as described in the legend to Fig. 2B. Note that the protected fragments representing readthrough transcripts are split into several bands due to the deletion of GB1 overlapping with the protection probe.