FIG. 6.
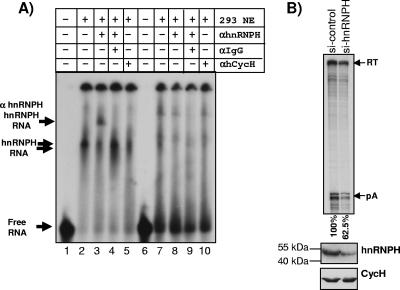
A) hnRNPH can interact with in vitro-transcribed GB1 sequences. Shown is an RNA band shift using in vitro-transcribed radiolabeled GB1 RNA (lanes 1 to 5) or control RNA (lanes 6 to 10) and HEK293 nuclear extract (NE). The table above the gel indicates addition (+) or omission (−) of components to the incubation mix: lane 1, no NE added; lanes 2 to 5 and 7 to 10, 0.25 μl nuclear extract added; lanes 3, 4, 8, and 9, 2 μl anti-hnRNPH antibody (αhnRNPH) added; lanes 4 and 9, anti-IgG antibody (αIgG) added; and lanes 5 and 10, 2 μl control anti-human cyclin H antibody (αhCycH) added. Free RNA and the formed complexes RNA-hnRNPH and RNA-hnRNPH-αhnRNPH are indicated by arrows on the left side. B) Upper panel, RNase protection of siRNA-treated HEK293 cells transiently transfected with the MC1R wild-type plasmid. si-control indicates HEK293 cells transfected with nontargeting siRNAs. si-hnRNPH represents HEK293 cells transfected with siRNAs targeting hnRNPH. Quantitation was performed using a Fuji phosphorimager, and the cleavage efficiency was calculated as the pA/RT ratio. Relative values of the processing activity are indicated below the gel and represent mean values of two independent experiments (62% and 63%). Lower panel, Western blot analysis of siRNA-treated HEK293 cells. hnRNPH, Western blot using an anti-hnRNPH-specific antibody; CycH, Western blot using a cyclin H-specific antibody.