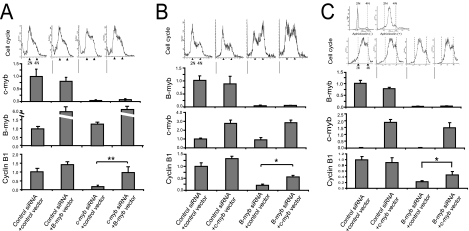
FIG. 3.
c-Myb substantially rescues cyclin B1 expression and G2/M transition in K562 and HCT116 cells. (A) K562 cells were transfected with either pcDNA3 (control vector) or pcDNA-B-myb (B-myb vector) expression constructs and either control or c-myb-targeted siRNA as described in Materials and Methods. Forty-eight hours posttransfection, a cell cycle analysis was conducted (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). For cell cycle analyses, the graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the B-myb vector, (iii) c-myb siRNA and the control vector, and (iv) c-myb siRNA and the B-myb vector, respectively. The combinations of the expression vector and siRNA that were transfected into K562 cells are indicated below the respective bar graphs. **, P < 0.01. (B) K562 cells were transfected with either the pcDNA3 (control vector) or the pcDNA-c-myb (c-myb vector) expression construct and either control or B-myb-targeted siRNA as described in Materials and Methods. Forty-eight hours posttransfection, a cell cycle analysis was performed (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). For cell cycle analyses, the graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the c-myb vector, (iii) B-myb siRNA and the control vector, and (iv) B-myb siRNA and the c-myb vector, respectively. The combinations of the expression vector and siRNA that were transfected into K562 cells are indicated below the respective bar graphs. *, P < 0.05. (C) HCT-116 cells transfected with either the pcDNA3 (control vector) or the pcDNA-c-myb (c-myb vector) expression construct and either control or B-myb-targeted siRNA as described in Materials and Methods. The cells were cultured in medium supplemented with aphidicolin for 24 h to synchronize the cells. The aphidicolin was removed from the medium, and the cells were then cultured for an additional 12 h. Thirty-six hours posttransfection, cell cycle analysis was conducted (four graphs above the bar graphs) and RNA was isolated, reverse transcribed, and amplified by real-time PCR with primers specific to c-myb, B-myb, and cyclin B1 (bar graphs). The combinations of the expression vector and siRNA that were transfected into HCT116 cells are indicated below the respective bar graphs. *, P < 0.05. The six graphs above the bar graphs correspond to cell cycle analyses. The two cell cycle analysis graphs above the four cell cycle analysis graphs correspond to subconfluent HCT116 cells in the absence (top left) and presence (top right) of aphidicolin 24 h prior to transfection. The bottom four cell cycle analysis graphs, from left to right, correspond to (i) control siRNA and the control vector, (ii) control siRNA and the c-myb vector, (iii) B-myb siRNA and the control vector, and (iv) B-myb siRNA and the c-myb vector, respectively. For all of the real-time PCR experiments, the expression of c-myb, B-myb, and cyclin B1 was normalized to that of GAPDH. The gene expression in the control cells for all of the bar graphs was then adjusted to 1.0, except for the c-myb bar graph with HCT-116 cells (C). For this graph (C), the relative expression of c-myb expression in control HCT116 cells was compared to c-myb expression in control K562 cells (B).