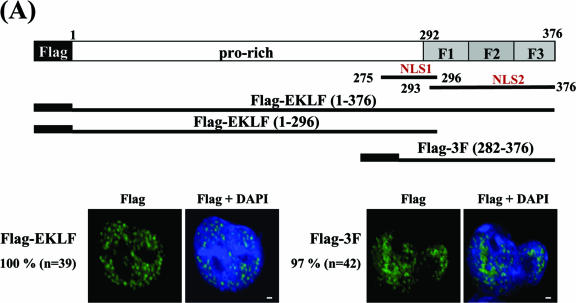
FIG. 7.
Mapping of EKLF domains required for DMSO-induced nuclear import of EKLF in MEL cells. (A) The subcellular locations of transiently expressed Flag-EKLF and Flag-3F in K562 cells. Top, schematic representation of the domain organization of EKLF. The proline-rich (pro-rich) region and the three-zinc-finger region (aa 292 to 376) are indicated. Shown below the EKLF map are the inserts of plasmid constructs pFlag-EKLF, pFlag-EKLF (1-296), and pFlag-3F used to transfect K562 and MEL cells. The subcellular locations of Flag-EKLF and Flag-3F in the transfected K562 cells were detected by immunostaining with anti-Flag. In either case, nearly all of the proteins were located in the nucleus. (B) The subcellular locations of transiently expressed Flag, Flag-EKLF, Flag-EKLF (1-296), and Flag-3F in MEL cells. Again, the locations of the exogenous proteins in the transfected MEL cells, before and after DMSO induction, were identified by immunostaining using anti-Flag as the probe. The nuclei were identified by DAPI staining. MetaMorph software (Meta Imaging Series version 6.1; Universal Imaging Corporation) was used to quantify the relative amounts of transiently expressed, Flag-tagged proteins in the nucleus and cytosol. For each experimental set, the percentage value (%) indicates the average fraction per cell of the corresponding exogenous protein located in the nucleus. The comparisons of the amounts of transiently expressed proteins in MEL cells before (white bars) and after (black bars) DMSO induction are shown schematically in the histogram for Flag, Flag-EKLF, Flag-EKLF (1-296), and Flag-3F. Note that transfected HA-EKLF behaved similarly to Flag-EKLF in MEL cells during DMSO induction (Shyu and Shen, unpublished results).