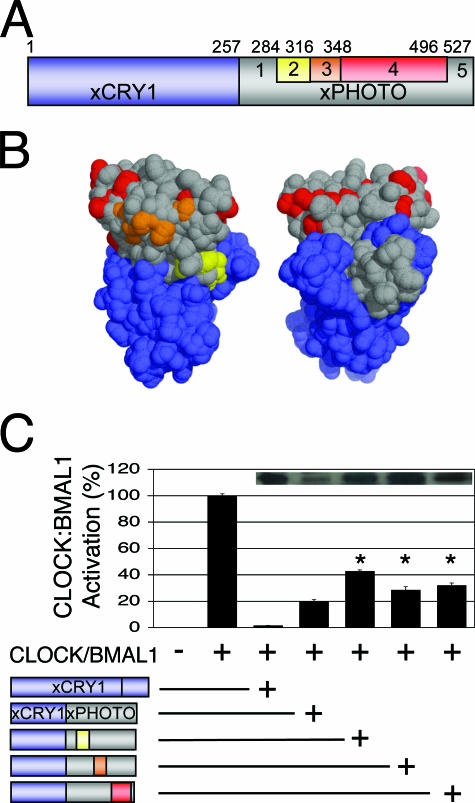
FIG. 5.
Replacing portions of the xPHOTO portion of xCRY1/xPHOTO with xCRY1 residues does not rescue repression. (A) Schematic diagram of the primary structure of the xCRY1/xPHOTO chimera (xCRY1 moiety, blue; xPHOTO moiety, gray), highlighting portions of the xPHOTO moiety that were replaced with the equivalent portions of xCRY1 to generate further chimeras: xCRY1/P1,C2,P3-5; xCRY1/P1-2,C3,P4-5; and xCRY1/P1-3,C4,P5 (yellow, red, and orange, respectively). (B) Homology model of the xCRY1 PHR generated using the SWISS-MODEL protein homology-modeling server (7) based on the structure of A. nidulans photolyase (PDB code 1OWL) (9) and visualized as a solvent-accessible surface using PyMOL (http://www.pymol.org) (4). This model displays residues that are from xCRY1 (blue) and xPHOTO (gray) in all xCRY/xPHOTO chimeras and also shows xCRY1 residues in the rescue chimeras that are conserved in vertebrate repressive-type CRYs but not xPHOTO (yellow, orange, and red). (C) Luciferase assay, carried out as described for Fig. 2. Full-length xCRY1 or xCRY1/xPHOTO chimeras were added as indicated (represented schematically). Repression by rescue chimeras was significantly worse than that by the original chimera (P < 0.01). Inset, Western blotting of FLAG-tagged xCRY1 and xCRY/xPHOTO chimeras, performed as described for Fig. 3. Rescue chimeras were expressed at levels similar to those of full-length xCRY1 and higher than those of the original chimera.