Abstract
The tumor suppressor p53 can induce apoptosis by activating gene expression in the nucleus, or by directly permeabilizing mitochondria in the cytoplasm. It has been shown that PUMA, a downstream target of p53 and a BH3-only Bcl-2 family member, plays an essential role in apoptosis induced by both nuclear and cytoplasmic p53. To understand how PUMA does so, we used homologous recombination to delete the binding sites of p53 in the promoter of PUMA in human colorectal cancer cells. As a result, the induction of PUMA and apoptosis in response to p53 and DNA-damaging agents were abrogated. Transcription coactivator recruitment and histone modifications in the PUMA promoter were suppressed. However, induction of PUMA and apoptosis in response to non-DNA-damaging stimuli were unaffected. These results indicate that the binding of nuclear p53 to the specific sites within the PUMA promoter is essential for its ability to induce apoptosis and is likely to be required for its tumor suppressive capacity.
Keywords: transcription, promoter, mitochondria, caspases, p53-binding sites
The tumor suppressor and transcription factor p53 binds to specific DNA sequences in promoter regions of its target genes after DNA damage (1). Activation of these genes by p53 triggers apoptosis, cell cycle arrest, DNA repair, and other responses (2). The consensus binding site of p53 was defined as two copies of the 10-bp motif (5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′) separated by a 0- to 13-bp spacer (3). It has been estimated that there are at least several hundred p53-binding sites in the human genome (4, 5). Although substantial evidence indicates that sequence-specific transcriptional activation by p53 is essential for its tumor-suppressor activity as well as its function in apoptosis (2, 6), it is unclear whether particular p53-binding sites are indispensable. It appears that every p53 target gene can be activated through p53-independent mechanisms. Furthermore, a number of recent studies have demonstrated that p53 can also induce apoptosis through a mechanism that does not depend on transactivation but, instead, involves translocation of p53 to the cytoplasm, where it directly activates proapoptotic Bcl-2 family proteins to permeabilize mitochondria (7, 8).
PUMA is a downstream target of p53 and a BH3-only Bcl-2 family member (9, 10). There are two p53-binding sites in the promoter region of PUMA, both of which can be directly bound and transactivated by p53 in vitro (9–11). Previous studies have demonstrated that PUMA plays an essential role in p53 and DNA damage-induced apoptosis in vitro and in vivo. Targeted deletion of PUMA in human colorectal cancer cells abrogates apoptosis induced by p53, DNA-damaging agents, γ-irradiation, and hypoxia (12). Knockout of PUMA in mice recapitulates several apoptotic deficiencies observed in p53-knockout mice (13, 14). PUMA also plays a role in p53-independent apoptosis induced by several stimuli, such as serum starvation and kinase inhibitors (14, 15). In addition to p53, other transcription factors, including the p53 homologue p73 and the FOXO family member FOXO3a, have been shown to activate PUMA expression (16, 17).
It has been shown that PUMA mediates apoptosis induced by both nuclear and cytoplasmic p53. Nuclear p53 activates the expression of PUMA, which in turn binds to antiapoptotic proteins Bcl-2 and Bcl- XL through its BH3 domain (9, 12), and relieves their inhibition on the proapoptotic Bcl-2 family protein, Bax (18). On the other hand, PUMA can also displace the cytoplasmic p53 from Bcl-XL, allowing p53 to directly activate Bax and induce mitochondrial permeabilization (19). Subsequently, mitochondrial apoptogenic proteins, including cytochrome C, SMAC, and AIF, are released into cytosol, leading to caspase activation and eventual cell death (9, 12).
To determine how PUMA mediates two seemingly distinct mechanisms of p53-induced apoptosis, we investigated whether the p53-binding sites in the PUMA promoter are necessary for PUMA induction and apoptosis in response to DNA damage.
Results
Targeted Deletion of the p53-Binding Sites in the PUMA Promoter.
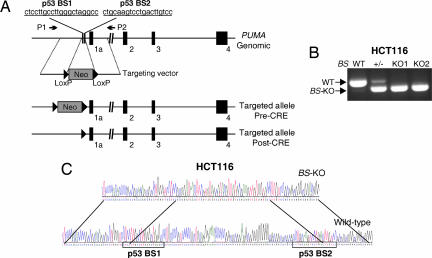
There are two sites that match the previously defined consensus binding site of p53 in the PUMA promoter region (Fig. 1A). Both sites can be bound by p53 and are required for p53 transactivation in reporter assays (9–11). These sites are not transcribed, therefore not amenable to RNA-based manipulation, such as RNAi. To study their functional role, we used homologous recombination to delete a 92-bp sequence including these sites in HCT116 colorectal cancer cells, which contain WT p53 and have been used as a model for studying apoptosis induced by endogenous p53 (20). Bioinformatic analysis indicated that the deleted region is devoid of binding sites of other transcription factors (data not shown). Knockout vectors were constructed by using a previously described recombinant adeno-associated virus (rAAV) system (Fig. 1A) (21). After two rounds of homologous recombination, HCT116 clones with the expected deletion were identified by PCR amplification of the corresponding genomic regions (Fig. 1B). DNA sequencing confirmed that both copies of the p53-binding sites (BS) were deleted in these clones (Fig. 1C).
Fig. 1.
Targeted deletion of the p53-binding sites in the PUMA promoter in HCT116 cells. (A) PUMA genomic locus and the targeting construct. The targeting construct consists of two homologous arms and the neomycin-resistance gene (Neo). Boxes 1a–4 represented exons 1a–4 of PUMA. Homologous recombination resulted in a deletion of 92 base pairs, including a part of both p53-binding sites (p53 BS1 and BS2). The same construct was used in the second round of gene targeting after the Neo, flanked by two LoxP sites, was excised from the heterozygous cells by Cre recombinase. The positions of the primers (P1 and P2) for PCR screening were indicated. (B) Identifying BS-KO clones. PCR was used to analyze HCT116 clones with different p53-binding site (BS) genotypes after removal of Neo. WT and the recombinant (BS-KO) alleles were indicated. (C) DNA sequences of the genomic region containing the p53-binding sites in the parental and BS-KO HCT116 cells. PCR and subsequent sequencing were performed after Neo was excised from both alleles. Two p53-binding sites and overlapping sequences between the WT and BS-KO alleles were indicated.
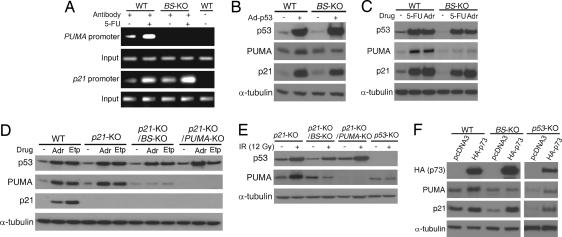
We first examined whether endogenous p53 is still able to bind to the PUMA promoter in the absence of the p53-binding sites. ChIP was used to analyze the binding of p53 to the PUMA promoter in cells treated with 5-fluorouracil (5-FU), a chemotherapeutic drug that induces p53-dependent apoptosis (22). In response to 5-FU, the binding of p53 to the PUMA promoter was significantly enhanced in the parental HCT116 cells, but undetectable in the p53-binding site-knockout (BS-KO) cells (Fig. 2A). In contrast, 5-FU-induced binding of p53 to the promoter of another p53 downstream target gene p21 was observed in both the parental and BS-KO cells (Fig. 2A). These results indicated that the specific binding of p53 to the PUMA promoter depends entirely on these two sites.
Fig. 2.
Induction of PUMA by DNA-damaging agents requires direct binding of p53 to the PUMA promoter. (A) The binding of p53 to the PUMA promoter. WT and BS-KO HCT116 cells were treated with 5-FU for 12 h. The bindings of p53 to the PUMA and p21 promoters were analyzed by ChIP, followed by PCR. Amplified region in the PUMA promoter is 5′ to the deleted region in the BS-KO cells. Amplified region in the p21 promoter is across the p53-binding sites. (B–F) Isogenic HCT116 cells with different p21, PUMA, BS, and p53 genotypes were subjected to the indicated treatments. Expression of p53, PUMA, and p21 was analyzed by Western blotting. α-Tubulin was used as the loading control. (B) Cells were infected with an adenovirus expressing p53 (Ad-p53) for 24 h. (C) Cells were treated with 5-FU (50 μg/ml) or adriamycin (Adr; 0.4 μg/ml) for 24 h. (D) Cells were treated with adriamycin (Adr; 0.4 μg/ml) or etoposide (Etp; 40 μM) for 24 h. (E) Cells were exposed to γ-irradiation (IR; 12 Gy), and cell lysates were prepared 24 h after the exposure. (F) Cells were transfected with HA-tagged p73 (HA-p73) or the control empty pCDNA3 vector. Cell lysates were prepared 24 h after transfection.
PUMA Induction in Response to DNA Damage Was Abrogated Without the p53-Binding Sites.
Next, we examined whether induction of PUMA by p53 requires the p53-binding sites. After infecting cells with an adenovirus expressing p53 (Ad-p53), we found that induction of PUMA, but not that of p21, in response to p53 overexpression was abolished in the BS-KO cells (Fig. 2B). To test whether PUMA induction by endogenous p53 requires the p53-binding sites, we treated parental and BS-KO HCT116 cells with 5-FU and the DNA-damaging agent adriamycin. Analysis of PUMA expression by Western blotting revealed that PUMA was induced by 5-FU and adriamycin in the parental cells, but not in the BS-KO cells (Fig. 2C). In contrast, the induction of p53 and p21 by these agents was not affected in the BS-KO cells (Fig. 2C). It has been shown that parental HCT116 cells undergo cell cycle arrest in response to p53 and DNA damage, whereas p21-deficient (p21-KO) HCT116 cells are subject to apoptosis induced by p53 and DNA damage (20). To study the role of the p53-binding sites in apoptosis, we deleted p21 in the BS-KO HCT116 cells (Fig. 2D). We found that the induction of PUMA by DNA-damaging agents adriamycin, etoposide, or γ-irradiation (IR, 12Gy) was abrogated in the p21 and BS double-knockout (p21-KO/BS-KO) cells (Fig. 2 D and E). The basal level of PUMA expression in the BS-KO cells remained unchanged after the treatment and was similar to that in the untreated parental cells, suggesting that the induction, rather than the basal expression of PUMA, depends on the binding of p53 to the PUMA promoter (Fig. 2 B–E). Interestingly, the induction of PUMA by the p53 homologue p73, which also plays a role in DNA damage-induced apoptosis (23), was abrogated in the BS-KO cells but unaffected in the isogenic p53-knockout (p53-KO) cells (Fig. 2F). These results indicated that induction of PUMA by p53 or p73 requires the p53-binding sites in the PUMA promoter.
p53-Binding Sites Are Necessary for DNA Damage-Induced Transcription Coactivator Recruitment and Histone Modification.
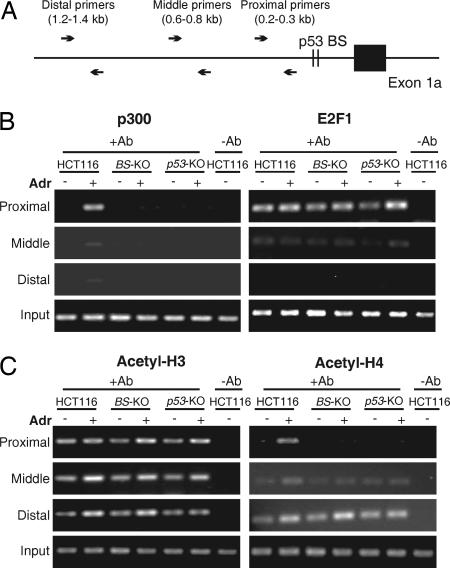
p53 can recruit transcription coactivators and trigger chromatin conformation changes to transcriptionally activate its target genes after DNA damage (24, 25). To test whether these events depend on the binding of p53 to the PUMA promoter, ChIP was used to analyze the binding of p300, a transcription coactivator that can be recruited by p53 in response to DNA damage (26). Adriamycin-induced binding of p300 to the PUMA promoter was found to be abolished in the BS-KO and p53-KO cells, suggesting that recruitment of p300 to the PUMA promoter by p53 occurs after the binding of p53 to this region (Fig. 3 A and B). In contrast, the binding of E2F1, a transcription factor that mediates the apoptotic function of p53 (27), to the PUMA promoter was unaltered in the BS-KO and p53-KO cells (Fig. 3B). ChIP was also used to analyze chromatin modifications in the PUMA promoter, including acetylation of histones H3 and H4. Histone H4 acetylation in the PUMA promoter, but not that of H3, was inhibited in the BS-KO and p53-KO cells after adriamycin treatment. However, these chromatin modifications were not affected in regions several hundred bp away from the p53-binding sites (Fig. 3C). These results suggested that the binding of p53 to specific DNA sequences is necessary for the recruitment of transcription coactivators to induce local chromatin modifications for transcription initiation at the PUMA promoter.
Fig. 3.
Recruitment of p300 and acetylation of histone H4 after DNA damage depend on the binding of p53 to the PUMA promoter. Parental, BS-KO, and p53-KO HCT116 cells were treated with adriamycin (Adr, 0.4 μg/ml) for 12 h. Chromatin complexes were cross-linked and isolated for ChIP analysis. (A) Map of the PCR primers for ChIP analysis. The positions of the primers that amplify a proximal, middle, or distal region relative to the p53-binding sites were indicated. (B) Bindings of p300 and E2F1 to the PUMA promoter. Immunoprecipitation (IP) was performed by using antibodies specific for p300 and E2F1. PCR was performed to quantify the amount of p300 and E2F1 bound to the indicated regions within the PUMA promoter. (C) Acetylation of histones H3 and H4. Antibodies specific for the acetyl-H3 and acetyl-H4 were used for ChIP. For “-Ab” controls, normal mouse or rabbit IgG was used. “Input” represented 1% of the starting materials for IP amplified by the proximal primers.
Apoptosis Induced by p53 or DNA Damage Was Abrogated Without the Binding of p53 to the PUMA Promoter.
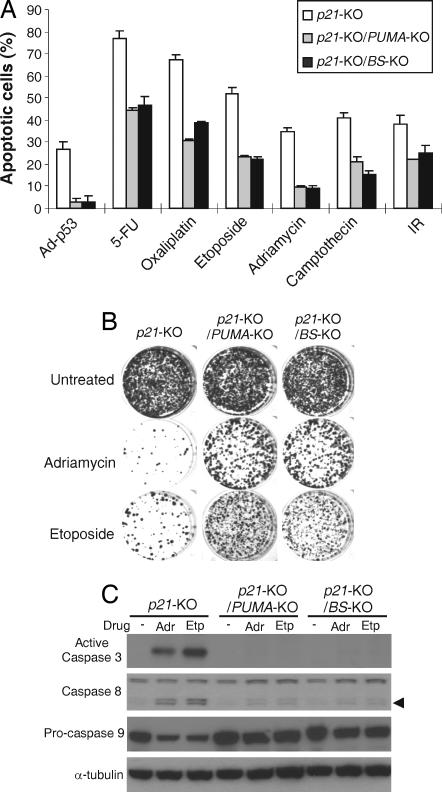
We then investigated whether PUMA-mediated apoptosis depends on the p53-binding sites. Our previous studies demonstrated that p21 and PUMA double-knockout cells (p21-KO/PUMA-KO) are deficient in apoptosis induced by p53 and DNA damage (12). Therefore, isogenic HCT116 cells, including p21-KO, p21-KO/PUMA-KO, and p21-KO/BS-KO cells, were compared for their response to p53 and DNA damage. Remarkably, apoptosis in response to Ad-p53 infection determined by nuclear staining was almost completely inhibited in the p21-KO/BS-KO cells as in the p21-KO/PUMA-KO cells (Fig. 4A). Apoptosis induced by DNA-damaging agents, including 5-FU, adriamycin, etoposide, oxaliplatin, camptothecin, and IR, was reduced to a similar extent in the p21-KO/BS-KO cells as in the p21-KO/PUMA-KO cells [Fig. 4A and supporting information (SI) Fig. 7). Analysis of apoptosis by Annexin V/propidium iodide staining, and measurement of long-term cell viability by colony formation assays, both confirmed that p21-KO/BS-KO cells were as resistant as the p21-KO/PUMA-KO cells to p53 and DNA-damaging agents (Fig. 4B and SI Fig. 7). Furthermore, DNA damage-induced caspase activation and release of cytochrome C from the mitochondria, which are characteristics of PUMA-induced apoptosis (9), were similarly inhibited in the p21-KO/BS-KO and the p21-KO/PUMA-KO cells (Fig. 4C and data not shown). Together, these results demonstrated that the binding of nuclear p53 to the PUMA promoter is required for PUMA-mediated caspase activation and apoptosis induced by DNA damage and p53.
Fig. 4.
PUMA-dependent apoptosis induced by p53 and DNA-damaging agents was abrogated in the BS-KO cells. HCT116 cells with the indicated genotypes were infected with Ad-p53 or treated with DNA-damaging agents, including 5-FU (50 μg/ml), oxaliplatin (50 μM), etoposide (40 μM), adriamycin (0.4 μg/ml), camptothecin (100 nM), or γ-irradiation (IR; 12 Gy). (A) Apoptosis determined by nuclear staining. After the indicated treatments for 48 h, attached and floating cells were fixed and analyzed by fluorescence microscopy after Hoechst 33258 staining. Those with fragmented and condensed nuclei were counted as apoptotic cells. (B) Colony formation assay for long-term cells survival. Approximately 500 cells treated with adriamycin (0.1 μg/ml) or etoposide (5 M) for 48 h were plated into 12-well plates. Colonies were visualized by crystal violet staining 2 weeks later. (C) Caspase activation. After adriamycin (Adr) and etoposide (Etp) treatments for 48 h, caspases 3, 8, and 9 were analyzed by Western blotting. The arrow indicated cleaved caspases. α-Tubulin was used as the loading control.
PUMA Induction and Apoptosis in Response to Non-DNA-Damaging Agents Do Not Depend on the Binding of p53 to the PUMA Promoter.
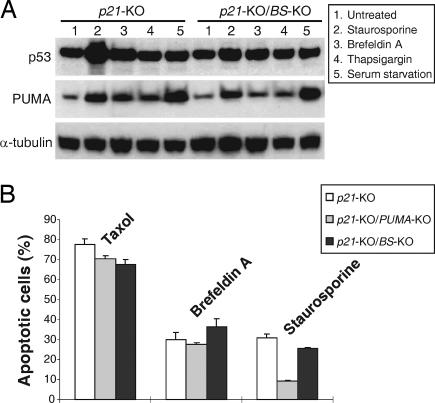
Because PUMA can be activated by non-DNA-damaging agents through p53-independent mechanisms (15), we also determined whether the induction of PUMA and apoptosis by these agents depend on the p53-binding sites. Kinase inhibitor staurosporine, endoplasmic reticulum (ER) stress inducers brefeldin A and thapsigargin, and serum starvation significantly induced PUMA in the p21-KO and p21-KO/BS-KO cells, suggesting that PUMA induction by these agents does not depend on the p53-binding sites (Fig. 5A). Staurosporine-induced apoptosis was suppressed in the p21-KO/PUMA-KO cells but not significantly altered in the p21-KO/BS-KO cells (Fig. 5B and SI Fig. 8), suggesting that staurosporine-induced apoptosis requires PUMA but does not rely on the binding of p53 to the PUMA promoter. The p53-binding sites and PUMA were not required for apoptosis induced by other non-DNA-damaging agents, such as taxol and brefeldin A (Fig. 5B). These results indicated that the binding of p53 to the PUMA promoter is dispensable for PUMA activation and apoptosis induced by non-DNA-damaging agents.
Fig. 5.
Induction of PUMA and apoptosis by non-DNA-damaging agents does not depend on the binding of p53 to the PUMA promoter. Cells with the indicated p21, PUMA, and BS genotypes were treated with non-DNA-damaging agents, including microtubule-targeting agent taxol (5 nM), kinase inhibitor staurosporine (60 nM), ER stress-inducers brefeldin A (10 nM) and thapsigargin (12 nM), or serum starvation. (A) Activation of PUMA. After the indicated treatments for 24 h, PUMA, p53, and α-tubulin were analyzed by Western blotting. (B) Apoptosis determined by nuclear staining. After the treatments for 48 h, apoptotic cells were counted after nuclear staining.
The Binding of p53 to the PUMA Promoter Is Necessary for p53-Induced Apoptosis in DLD1 Cells.
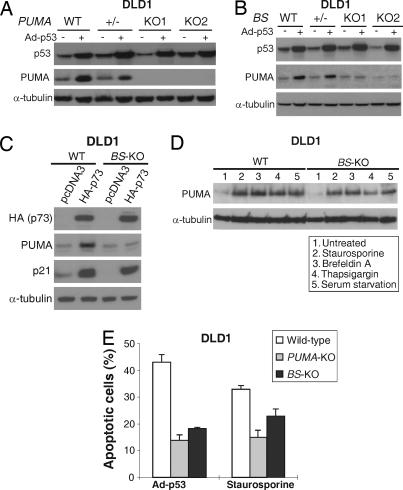
To rule out the possibility that our observations were due to the specific genetic background in HCT116 cells, we analyzed another colorectal cancer cell line DLD1. This cell line is p53-deficient and undergoes apoptosis in response to exogenous p53 expression. We first deleted the coding region of PUMA in DLD1 cells by homologous recombination using the rAAV system (SI Fig. 9A). Western blotting confirmed that there is no PUMA expression in the PUMA-KO DLD1 cells (Fig. 6A). The p53-binding sites in the PUMA promoter were also deleted in DLD1 cells by using the same construct for targeting HCT116 cells (SI Fig. 9B). The induction of PUMA by p53 or p73 was completely abolished in the p53 BS-KO DLD1 cells (Fig. 6 B and C), whereas the induction of PUMA by non-DNA-damaging stimuli was not affected (Fig. 6D). Furthermore, apoptosis induced by p53, but not that induced by staurosporine, was abrogated to a similar extent in the BS-KO DLD1 cells as in the PUMA-KO DLD1 cells (Fig. 6E). These results confirmed that induction of PUMA and apoptosis by p53 and DNA-damaging agents, rather than non-DNA-damaging agents, strictly depend on the binding of nuclear p53 to the PUMA promoter.
Fig. 6.
Targeted deletion of PUMA or p53-binding sites abrogated p53-induced apoptosis in DLD1 cells. (A) Targeting PUMA in DLD1 cells. Cells with indicated PUMA genotypes were infected by Ad-p53 for 24 h. p53, PUMA and α-tubulin were analyzed by Western blotting. (B) Targeting the BS in DLD1 cells. DLD1 cells with the indicated BS genotypes were infected with Ad-p53 for 24 h. p53, PUMA and α-tubulin were analyzed by Western blotting. (C) Induction of PUMA by p73. DLD1 cells with the indicated genotypes were transfected with HA-tagged p73 (HA-p73) or the control empty pCDNA3 vector. The expression of the indicated proteins at 24 h after transfection was analyzed by Western blotting. (D) Induction of PUMA by non-DNA-damaging agents. WT and BS-KO DLD1 cells were treated with the indicated non-DNA-damaging agents for 24 h. PUMA, p53, and α-tubulin were analyzed by Western blotting. (E) Apoptosis in the PUMA-KO and BS-KO DLD1 cells. WT, PUMA-KO, and BS-KO DLD1 cells were infected with Ad-p53 or treated with staurosporine. Apoptosis was analyzed by nuclear staining 48 h after the treatment.
Discussion
Our results demonstrated that transcriptional activation of PUMA, which requires the direct binding of nuclear p53 to the p53-binding sites in the PUMA promoter, accounts for nearly all of DNA damage-induced PUMA activation and PUMA-mediated apoptosis. Events driving transcription initiation, such as recruitment of transcription coactivators and chromatin modifications, can occur only after p53 occupies the PUMA promoter. These results suggested a direct, specific, and essential role of the p53-binding sites in these processes. In conjunction with the data indicating an essential role of PUMA in DNA damage-induced and p53-mediated apoptosis (12–14), these results strongly suggested that transcriptional activation of PUMA by nuclear p53 is indispensable for initiation of DNA damage-induced apoptosis.
The previously reported transcription-independent function of p53 in apoptosis induction may be reconciled by several possibilities. Only certain stimuli may rely on the transcription-independent function of p53 to induce apoptosis. For example, apoptosis induced by UV radiation in HCT116 cells was found to be mediated by PUMA and involve cytoplasmic p53 (19). The p53-binding site-knockout cell lines generated in this study will be useful for determining the contributions of nuclear and cytoplasmic p53 in apoptosis induced by different stimuli. It is also possible that accumulation of PUMA through p53-medaited transcriptional activation serves as a prerequisite for the displacement of cytoplasmic p53 from antiapoptotic proteins by PUMA. Without induction, the endogenous PUMA may not be sufficient to displace cytoplasmic p53 from antiapoptotic proteins to allow permeabilization of mitochondrial membrane. Future experiments are necessary to determine how p53-mediated nuclear and cytoplasmic events are coordinated to trigger apoptotic initiation.
PUMA appears to be a converging molecule through which a variety of stimuli induce apoptosis. It is specifically activated by p53 or p73 in response to DNA damage. Induction of PUMA by p73 is p53-independent and requires the p53-binding sites. However, p53 seems to play a major role in regulating endogenous PUMA expression, because the induction of PUMA by DNA-damaging agents strictly depends on the presence of functional p53 (9, 10, 28). The p53-PUMA pathway can be modulated by other proteins. For example, a recent study showed that the transcription repressor Slug antagonizes p53-mediated apoptosis by repressing PUMA in hematopoietic progenitor cells (29). Certain non-DNA-damaging agents, such as the kinase inhibitor staurosporine, elicit PUMA-dependent apoptosis that does not require the p53-binding sites. Activation of PUMA by these agents is likely to be mediated by binding sites of other transcription factors in the PUMA promoter. For example, a recent study found that FOXO3a, a member of the FOXO family of transcription factors, up-regulates PUMA expression in response to cytokine or growth factor deprivation in a p53-independent manner (17). Further studies are necessary to pinpoint the transcription factors and DNA elements that drive PUMA up-regulation in response to p53-independent apoptotic stimuli.
Materials and Methods
Cell Culture.
Human colorectal cancer cell lines HCT116 and DLD1 and their derivative cell lines were cultured in McCoy's 5A modified media (Invitrogen, Carlsbad, CA) supplemented with 10% defined FBS (HyClone, Logan, UT), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were maintained at 37°C with 5% CO2. Lipofectamine 2000 (Invitrogen) was used to transfect cells according to the manufacture's instructions.
Targeting the p53-Binding Sites in the PUMA Promoter.
Gene targeting vectors were constructed by using the previously described rAAV system (21). Briefly, two homologous arms flanking the p53-binding sites, which are 1.2 kb and 1.15 kb, respectively, along with the neomycin-resistant gene cassette (Neo), were inserted between two NotI sites in the AAV shuttle vector pAAV-MCS (Stratagene, La Jolla, CA). Packaging of rAAV was performed by using the AAV Helper-Free System (Stratagene) according to the manufacturer's instructions.
HCT116 and DLD1 cells containing two copies of WT PUMA alleles were infected with rAAV and selected by G418 (0.4 mg/ml) for 3 weeks. G418-resistant clones were pooled and screened by PCR for targeting events by using different primer pairs listed in SI Table 1. To target the second allele, Neo flanked by two Lox P sites was excised from a heterozygous clone by infecting cells with an adenovirus expressing Cre recombinase (Ad-Cre) (12). The same targeting construct was used in the second round of gene targeting. After the second round, Neo was excised by Ad-Cre infection, and gene targeting was verified by genomic sequencing.
To generate cell lines deficient in both p53-binding sites and p21, p53-binding site-knockout cells were subjected to two rounds of gene targeting for p21 by using a previously described method (30). Homologous recombination at the p21 locus was identified by PCR and confirmed by immunoblotting. To knock out PUMA in DLD1 cells, rAAV targeting vector was constructed by using the previously described homologous arms for targeting PUMA (12). Knockout clones were identified and confirmed as described (12). The PCR primers used for generating knockout constructs and for identifying recombinant clones were listed in SI Table 2. The detailed procedures of gene targeting and PCR screening are available upon request.
Drug Treatment, Irradiation and Adenovirus Infection.
Cells were plated in 12-well plates at 20–30% density 24 h before treatment. For drug treatment, DMSO stock solutions of anticancer agents were diluted into appropriate concentrations with cell culture medium. The anticancer agents, including adriamycin, etoposide, camptothecin, oxaliplatin, 5-fluorouracil (5-FU), and taxol, were from Sigma (St. Louis, MO). Thapsigargin, (+)-brefeldin A and staurosporine were from EMD Biosciences (San Diego, CA). A Cs137 γ source was used for ionizing radiation. The adenoviruses expressing WT p53 (Ad-p53) were previously described (12).
Apoptosis Assays.
After the treatment, attached and floating cells were harvested at various time points. Apoptosis was analyzed by counting cells with condensed chromatin and micronucleation after nuclear staining with Hoechst 33258 as described (31). A minimum of 300 cells was analyzed each time. Cells were also stained by Annexin V Alexa Fluor 488 conjugate and propidium iodide (Invitrogen), and analyzed by flow cytometry. Long-term cell survival after drug treatment was determined by colony-formation assays. In brief, cells were plated at appropriate dilutions after the treatment, allowed to grow for 10–14 days, and stained by crystal violet (Sigma). All apoptosis measurements and colony formation assays were repeated at least three times. The mean ± SD are displayed in the figures.
Western Blotting and Antibodies.
Cell lysates were collected, and immunoblotting was performed as described (31). The antibodies used for Western blotting included rabbit polyclonal antibodies against PUMA (12), p53 (DO1), HA (Santa Cruz Biotechnology, Santa Cruz, CA), p21, α-tubulin (EMD Biosciences), active caspase 3 (Stressgen Bioreagents, Ann Arbor, MI), caspase 8, and caspase 9 (Cell Signaling Technology, Beverly, MA).
ChIP.
ChIP was performed by using the Chromatin Immunoprecipitation Assay kit (Upstate Biotechnology, Lake Placid, NY) according to manufacturer's instructions with minor modifications. Briefly, ≈2 × 106 cells were fixed with 1% formaldehyde and lysed in SDS lysis buffer. DNA in the cross-linked chromatin preparations was sheared to 200–1,000 bp by sonication. Samples were precleared with salmon sperm DNA/protein A agarose (50%) slurry. After addition of antibodies and fresh protein A agarose, the samples were incubated at 4°C overnight. Normal mouse or rabbit IgG were used as controls. Precipitated chromatin complexes were eluted by 500 μl of elution buffer (1% SDS, 0.1 M NaHCO3) for 30 min. Finally, the protein-DNA cross-links were reversed by overnight incubation with 100 μM NaCl at 65°C, and immunoprecipitated DNA was analyzed by PCR using primers listed in SI Table 3. Antibodies for ChIP included rabbit polyclonal antibodies against E2F1 and p300 (Santa Cruz Biotechnology), and monoclonal antibodies for acetylated histones H3 and H4 (Upstate Biotechnology).
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein (The Sidney Kimmel Comprehensive Cancer Center at Hopkins, Baltimore, MD) for providing the rAAV-knockout system, p21-KO and p53-KO cells; Dr. William G. Kaelin, Jr., (Dana–Farber Cancer Institute, Boston, MA) for p73 expression construct; and Drs. Richard A. Steinman and Shiyuan Cheng for careful reading and comments on the manuscript. This work was supported by National Institutes of Health Grant CA106348, grants from the Edward Mallinckrodt, Jr. Foundation (to L.Z.), and grants from the Flight Attendant Medical Research Institute and the Alliance for Cancer Gene Therapy (to J.Y.). L.Z. is a scholar of the General Motors Cancer Research Foundation and the V Foundation for Cancer Research.
Abbreviations
- PUMA
p53 up-regulated modulator of apoptosis
- SMAC
second mitochondria-derived activator of caspase
- AIF
apoptosis-inducing factor
- FOXO3A
Forkhead Box O3A
- BH3
Bcl-2 homology 3
- BS
p53-binding sites
- 5-FU
5-fluorouracil
- AAV
adeno-associated virus
- Ad
adenovirus.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700020104/DC1.
References
- 1.Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 4.Tokino T, Thiagalingam S, el-Deiry WS, Waldman T, Kinzler KW, Vogelstein B. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 5.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Zhang L. Biochem Biophys Res Commun. 2005;331:851–858. doi: 10.1016/j.bbrc.2005.03.189. [DOI] [PubMed] [Google Scholar]
- 7.Moll UM, Wolff S, Speidel D, Deppert W. Curr Opin Cell Biol. 2005;17:631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Chipuk JE, Maurer U, Green DR, Schuler M. Cancer Cell. 2003;4:371–381. doi: 10.1016/s1535-6108(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 10.Nakano K, Vousden KH. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 11.Kaeser MD, Iggo RD. Proc Natl Acad Sci USA. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A. Science. 2003;302:1036–1038. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 14.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Flemington C, Houghton AB, Gu Z, Zambetti GP, Lutz RJ, Zhu L, Chittenden T. Proc Natl Acad Sci USA. 2001;98:11318–11323. doi: 10.1073/pnas.201208798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 17.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, Erlacher M, Villunger A, Mak TW. J Exp Med. 2006;203:1657–1663. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ming L, Wang P, Bank A, Yu J, Zhang L. J Biol Chem. 2006;281:16034–16042. doi: 10.1074/jbc.M513587200. [DOI] [PubMed] [Google Scholar]
- 19.Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR. Science. 2005;309:1732–1735. doi: 10.1126/science.1114297. [DOI] [PubMed] [Google Scholar]
- 20.Polyak K, Waldman T, He T-C, Kinzler KW, Vogelstein B. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 21.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jost CA, Marin MC, Kaelin WG., Jr Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 24.Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 25.Coutts AS, La Thangue NB. Biochem Biophys Res Commun. 2005;331:778–785. doi: 10.1016/j.bbrc.2005.03.150. [DOI] [PubMed] [Google Scholar]
- 26.Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 27.Pan H, Yin C, Dyson NJ, Harlow E, Yamasaki L, Van Dyke T. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Yue W, Wu B, Zhang L. Clin Cancer Res. 2006;12:2928–2936. doi: 10.1158/1078-0432.CCR-05-2429. [DOI] [PubMed] [Google Scholar]
- 29.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Waldman T, Kinzler KW, Vogelstein B. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 31.Kohli M, Yu J, Seaman C, Bardelli A, Kinzler KW, Vogelstein B, Lengauer C, Zhang L. Proc Natl Acad Sci USA. 2004;101:16897–16902. doi: 10.1073/pnas.0403405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.