Fig. 4.
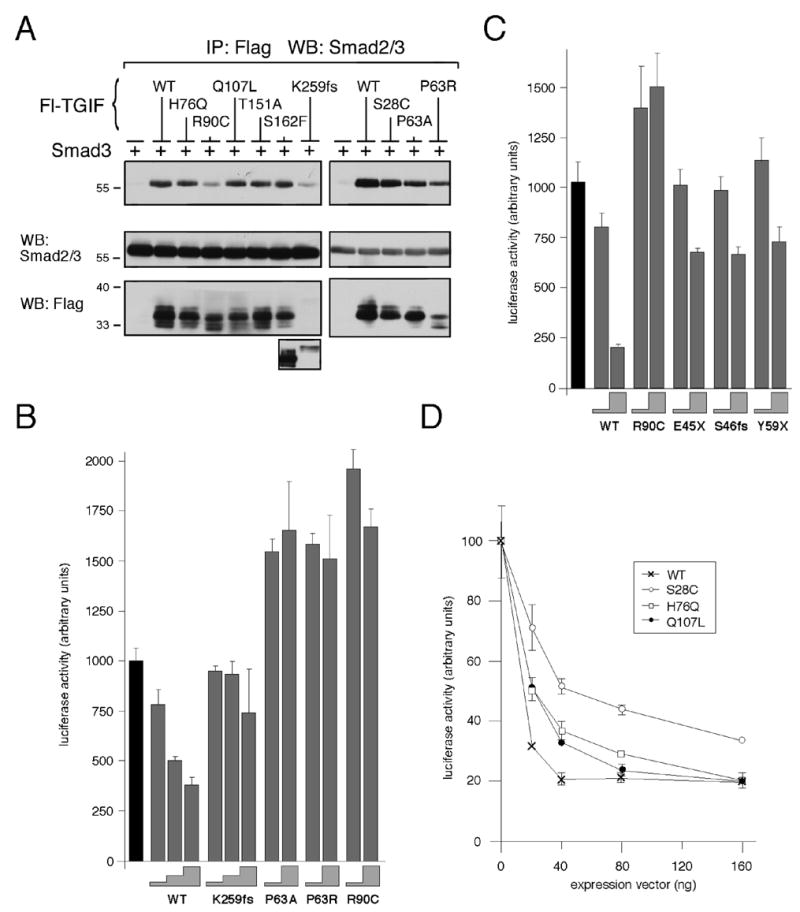
TGIF HPE mutants interact with Smad3 and affect repression of TGFβ-activated transcription. (A) COS1 cells were transfected with untagged Smad3 and Flag-TGIF wildtype or HPE mutants or control vector, as indicated. Protein complexes were isolated on anti-Flag agarose and analyzed by western blot for the presence of coprecipitating Smad3. A portion of the lysates was subjected to direct western blot analysis to monitor protein expression (below). Due to the low expression level of the K259fs construct, a longer exposure of the lysate western blot is shown below. (B, C and D) HepG2 cells were transfected with a TGFβ-responsive luciferase reporter, 3TP-lux, and increasing amounts of TGIF wildtype or HPE mutant expression constructs. All cells were treated with 100 pM TGFβ prior to analysis. Firefly luciferase activity was normalized to Renilla luciferase and is represented in arbitrary units, as the mean +/− s.d. of duplicate transfections. (B) HepG2 cells were transfected with 10 ng and 50, or 10, 25 and 50 ng of TGIF expression construct (the height of the grey steps indicate the amount of input TGIF DNA: 10, 25 or 50 ng). The black bar indicate the activity of the reporter alone, without TGIF (C) 10 or 100 ng of TGIF constructs were transfected (grey steps). (D) Dose-dependent repression of luciferase activity by TGIF is shown as a graph of luciferase activity in arbitrary units as the mean +/− s.d. of duplicate transfections as a function of the amount (in ng) of TGIF expression vector transfected.
