Abstract
PrPSc [abnormal disease-specific conformation of PrP (prion-related protein)] accumulates in prion-affected individuals in the form of amorphous aggregates. Limited proteolysis of PrPSc results in a protease-resistant core of PrPSc of molecular mass of 27–30 kDa (PrP27–30). Aggregated forms of PrP co-purify with prion infectivity, although infectivity does not always correlate with the presence of PrP27–30. This suggests that discrimination between PrPC (normal cellular PrP) and PrPSc by proteolysis may underestimate the repertoire and quantity of PrPSc subtypes. We have developed a CDI (conformation-dependent immunoassay) utilizing time-resolved fluorescence to study the conformers of disease-associated PrP in natural cases of sheep scrapie, without using PK (proteinase K) treatment to discriminate between PrPC and PrPSc. The capture-detector CDI utilizes N-terminal- and C-terminal-specific anti-PrP monoclonal antibodies that recognize regions of the prion protein differentially buried or exposed depending on the extent of denaturation of the molecule. PrPSc was precipitated from scrapie-infected brain stem and cerebellum tissue following sarkosyl extraction, with or without the use of sodium phosphotungstic acid, and native and denatured PrPSc detected by CDI. PrPSc was detectable in brain tissue from homozygous VRQ (V136 R154 Q171) and ARQ (A136 R154 Q171) scrapie-infected sheep brains. The highest levels of PrPSc were found in homozygous VRQ scrapie-infected brains. The quantity of PrPSc was significantly reduced, up to 90% in some cases, when samples were treated with PK prior to the CDI. Collectively, our results show that the level of PrPSc in brain samples from cases of natural scrapie display genotypic differences and that a significant amount of this material is PK-sensitive.
Keywords: conformation-dependent immunoassay, epitope, prion protein, proteinase K, proteolysis, transmissible spongiform encephalopathy
Abbreviations: AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride; ARQ, A136 R154 Q171; BCA, bicinchoninic acid; BSE, bovine spongiform encephalopathy; CDI, conformation-dependent immunoassay; CNS, central nervous system; cps, counts per second; GdnHCl, guanidine hydrochloride; mAb, monoclonal antibody; PK, proteinase K; PrP, prion-related protein; PrPC, normal cellular PrP; PrPSc, abnormal disease-specific conformation of PrP; TRF, time-resolved fluorescence; TSE, transmissible spongiform encephalopathy; VRQ, V136 R154 Q171
INTRODUCTION
Prion diseases, or TSEs (transmissible spongiform encephalopathies), are chronic neurodegenerative CNS (central nervous system) disorders of mammals. This group of invariably fatal conditions includes scrapie of sheep, BSE (bovine spongiform encephalopathy) of cattle, CWD (chronic wasting disease) of cervids and CJD (Creutzfeldt–Jakob disease) of humans. These diseases are characterized by the accumulation of PrPSc [abnormal disease-specific conformation of PrP (prion-related protein)], an abnormal isomer of the host protein PrPC (normal cellular PrP), in the brains, and in some cases peripheral tissues of affected individuals [1]. PrPC is a GPI (glycosylphosphatidylinositol) membrane-linked protein of molecular mass of 32–35 kDa [2]. Although the covalent structure of PrPC and that of PrPSc appear identical, the two isomers of PrP have significantly different biophysical properties. PrPC is soluble in detergent and readily digested by proteases, whereas PrPSc is insoluble in detergent and relatively resistant to proteolytic digestion, at least within the C-terminal region of the molecule [3]. Fourier-transform infrared spectroscopy has shown that PrPC is predominantly α-helical (42%) with little β-sheet (3%), whereas PrPSc has considerably more β-sheet content (43%) and a similar α-helical content (30%) [4,5]. These observations indicate that during conversion of PrPC into PrPSc, a major refolding event occurs that results in a more extensive β-sheet conformation. This conformational change would appear to be fundamental to prion propagation, and substantial evidence suggests that the infectious prion agent comprises solely proteinaceous material in the form of an abnormal isomer of PrP [6–8]. Different prion inocula, or strains, may be characterized by several criteria, including their biological properties, histopathology, and variations in the pattern of PrPSc deposition and the length of the disease incubation period following experimental inoculation [9].
PrPSc accumulates principally in the brain of prion-affected individuals in the form of amorphous aggregates. Detergent extraction and limited proteolysis of PrPSc result in fibrillar aggregates of a protease-resistant core that polymerize spontaneously into amyloid [10]. The protease-resistant core of PrPSc has a molecular mass of 27–30 kDa (PrP27–30) and typically lacks the N-terminal 67–70 amino acids of the normal prion protein [11]. Aggregated forms of PrP co-purify with prion infectivity, although infectivity does not always correlate with the presence of PrP27–30 [12–15]. This has led to concerns that limited digestion with proteases to discriminate between PrPC and PrPSc may lead to an underestimate of the repertoire of conformations of disease-associated PrP and their quantity within prion-infected tissues. Typically, the conformations of multimeric disease-associated PrP isoforms have been studied through the use of immunoassays that recognize regions of the protein that are differentially buried or exposed depending on the extent of denaturation of the molecule. Peretz et al. [16] showed that regions of PrP were buried in native PrPSc but solvent-exposed in PrPC. Epitopes buried in PrPSc showed a substantial increase in immunoreactivity after denaturation of the PrP molecule, whereas those that were solvent-exposed did not change in their reactivity. This approach has led to the development of the CDI (conformation-dependent immunoassay), which is capable of quantifying the concentration of PrPSc without the use of PK (Proteinase K) to discriminate between PrPC and the abnormal isomer of PrP [17]. The CDI revealed that PrPSc adopts both protease-resistant and -sensitive conformations. In addition, prion strains were found to differ from each other by the extent to which those epitopes that were buried under native conditions were increased upon partial denaturation of disease-associated PrP. In an alternative approach, Paramithiotis et al. [18] have shown that regions of PrPC that are not solvent-exposed are accessible to certain anti-PrP antibodies in PrPSc. These approaches have highlighted the utility of mAbs (monoclonal antibodies) to probe conformations of disease-associated PrP through the analysis of the accessibility of distinct PrP epitopes.
Scrapie in sheep is the archetypal prion disease, and polymorphisms in ovine PrP at amino acid residues 136, 154 and 171 are associated with variation in susceptibility to natural or typical scrapie. VRQ (V136 R154 Q171) or ARQ (A136 R154 Q171) animals show susceptibility to scrapie, whereas those that express ARR (A136 R154 R171) or AHQ (A136 H154 Q171) show resistance [19–22]. All three polymorphic sites are located within, or close to, that region of PrP that undergoes the major conformational change associated with conversion of PrPC into PrPSc during prion disease [23]. Polymorphisms in ovine PrP appear to affect the stability of the molecule and also its amyloidogenic potential [24–26]. These biophysical properties of ovine VRQ PrP correlate with the observation that homozygous VRQ sheep show the shortest incubation time for the development of experimental and natural forms of typical scrapie. Collectively, these observations have led to the hypothesis that differences in the metabolism of allelic variants of ovine PrP contribute to the mechanism(s) that determine susceptibility and resistance of sheep to natural scrapie [24,25]. These differences in metabolism may lead to a variation in accumulation of PrPSc and its subtypes in scrapie-infected sheep of different genotypes.
In the present study, we have developed a CDI based on the use of N-terminal- and C-terminal-specific anti-PrP mAbs, and TRF (time-resolved fluorescence), to investigate ovine PrPSc in scrapie-infected sheep brain samples. PrPSc is typically found at high levels in the brain stem and cerebellum of sheep infected with classical scrapie, and brain stem is the primary tissue used for routine diagnosis of scrapie. However, it has been reported that in cases of atypical scrapie the brain stem has limited PrPSc burden, while high levels may be found in the cerebellum [27]. Accordingly, in the study presented here both brain stem and cerebellum were investigated to ensure the utility of both tissues for immunodetection of ovine PrPSc by CDI. For the first time, we show that PrPSc from natural isolates of homozygous VRQ scrapie-infected sheep contain more PrPSc at terminal disease than homozygous ARQ sheep. Furthermore, we have shown that ovine disease-associated PrP comprises PK-sensitive and -resistant forms of PrPSc. Most of the PrPSc in both homozygous VRQ and ARQ scrapie-infected sheep brains was found to be PK-sensitive. These observations provide new and novel insights into the molecular characteristics of ovine PrPSc.
MATERIALS AND METHODS
Generation of ovine recombinant PrP
Full-length ovine recombinant ARQ and VRQ (amino acid residues 25–232) were generated as described previously [28]. PrP proteins were verified by MS to confirm the correct protein sequence and the presence of a disulphide bond. Oxidized and refolded recombinant PrP was stored at −80 °C.
Anti-PrP mAbs
The N-terminal-specific mAb FH11 [29,30] was purchased from the TSE Resource Centre (Compton, Berks., U.K.). The C-terminal-specific anti-PrP mAbs A516, V24 and V26 were generated from Prnpo/o mice immunized with ovine recombinant PrP [28]. Anti-PrP mAb 245 was generated from Prnpo/o mice immunized with copper-refolded full-length murine recombinant PrP (amino acid residues 23–231) [31]. Anti-PrP mAb 683 was generated from Prnpo/o mice immunized with a peptide of murine recombinant PrP (amino acid residues 161–231) [28]. Biotinylated mAbs were prepared for use as detector antibodies as follows. One milligram of each purified mAb was washed four times with 50 mM sodium bicarbonate buffer (pH 9.3) using YM-30 concentrators (Fisher UK, catalogue no. FDR-563-020L). The non-diffusible material was then labelled overnight at 4 °C using N,N-dimethylformamide (Sigma, catalogue no. D-4551) and biotinamidocaproate N-hydroxysuccinimide ester (Sigma, catalogue no. B-2643). Biotinylated antibodies were purified by adsorption on to YM-30 concentrator membranes followed by isolation using elution buffer (50 mM Tris/HCl, pH 7.8, 0.9% sodium chloride and 0.1% sodium azide, pH 7.8). The immunoreactivity of biotinylated mAbs was verified by direct ELISA using ovine recombinant PrP as antigen and the concentration of antibody was determined by BCA (bicinchoninic acid) assay (Pierce, Perbio Science, catalogue no. 23225) prior to storage at 4 °C.
Preparation of sheep brain tissues
Various ovine brain regions were prepared by two cycles of homogenization in PBS (pH 7.4) in a Bio-Rad TeSeE Precess 24 homogenizer and individual samples were diluted to 10% in PBS prior to being tested.
Protein quantification
Total protein concentration in ovine brain material, ovine recombinant PrP or anti-PrP mAb samples was measured using the BCA assay. Samples were initially diluted in PBS (pH 7.4), at 1:50, 1:100 and 1:200 dilutions plus a diluent-only control. A 2-fold dilution series of BSA (Sigma, catalogue no. A-3059) was prepared ranging from 800 μg/ml down to 0 μg/ml to act as a standard. Each dilution was dispensed into triplicate wells in a 96-well flat-bottomed plate (5 μl/well). An aliquot of 95 μl of BCA working reagent was added to each well and the plates were incubated for up to 30 min at 20 °C. Absorbance was read at 570 nm using a Bio-Rad 680 microplate reader. Protein concentration was estimated by calculation from the standard curve using Microplate Manager software (Bio-Rad).
Enrichment for disease-associated PrP
The 10% tissue homogenates were treated with or without protease inhibitors at final concentrations of 0.5 mM PMSF diluted in 100% ethanol, 2 μg/ml leupeptin diluted in distilled water and 2 μg/ml aprotinin diluted in distilled water. Some samples were treated with or without 1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride; Sigma, catalogue no. A-8456] diluted in distilled water instead of the other protease inhibitors listed above. Samples were centrifuged at 100 g for 1 min at 20 °C to remove gross debris and the supernatants were retained. A final concentration of 2% (v/v) sarkosyl in PBS (pH 7.4) was added to all samples and incubated for 10 min at 37 °C with shaking prior to the addition of MgCl2 at a final concentration of 1.15 mM and incubation for 30 min at 37 °C with constant shaking. In some cases, samples were treated with or without PK (Roche, catalogue no. 1000 144) at a final concentration of 16 μg/ml in PBS (pH 7.4) for 30 min at 37 °C with constant shaking (prior to the addition of any protease inhibitors). The enzymatic reactions were stopped by the addition of a final concentration of 1 mM AEBSF diluted in distilled water and samples were then treated with or without sodium phosphotungstic acid at a final concentration of 0.4% containing 17 mM MgCl2 diluted in PBS (pH 7.4) for 2 or 20 h at 37 °C with shaking. Samples were centrifuged at 21000 g for 30 min at 10 °C, the supernatants were discarded and the pellets were resuspended in 200 μl of 0.1% (v/v) sarkosyl in PBS (pH 7.4) and 200 μl of 250 mM EDTA in water (pH 8.0) to solubilize the magnesium salts. Samples were thoroughly mixed and either stored at 4 °C overnight and then centrifuged or centrifuged immediately at 21000 g for 15 min at 10 °C. The supernatants were discarded and the pellets were resuspended in 0 or 6 M GdnHCl (guanidine hydrochloride). All 6 M GdnHCl-treated samples were then heated to 80 °C for 5 min, cooled to 20 °C and then diluted as required.
CDI
For CDI, capture antibody (FH11 or A516) was routinely coated at 0.5 μg/well in triplicate in Nunc Maxisorp 96-well flat-bottomed plates overnight at 4 °C. Capture antibodies were diluted in coating buffer (0.01 M PBS, pH 7.4, containing 0.1% sodium azide). Excess antibody was removed and plates were washed four times with wash buffer (25× concentrated stock prepared as 500 mM Tris/HCl pH 7.7, 154 mM NaCl, 0.5% Tween 20 and 0.1% sodium azide diluted to 1× working concentration with distilled water just prior to use) and wells were blocked with block buffer consisting of 2% (w/v) BSA, and 0.05% sodium azide in PBS for 1.5 h at 20 °C with shaking. Plates were washed four times with wash buffer and the appropriate dilutions of ovine recombinant PrP protein or ovine tissue samples were diluted as required in assay buffer [50 mM Tris/HCl, pH 7.7, 154 mM NaCl, 0.02 mM diethylenetriaminepenta-acetic acid (Sigma, catalogue no. D-6518), 0.5% BSA, 0.1% sodium azide and 0.01% Tween 20]. Samples were incubated for 1 h at 20 °C with shaking. Plates were washed four times with wash buffer and the relevant biotinylated anti-PrP detector mAb was added at 50 ng/well diluted in assay buffer and incubated for 1 h at 20 °C with shaking. Plates were washed four times with wash buffer followed by incubation with Europium-labelled streptavidin (PerkinElmer, catalogue no. 1244-360) at 50 ng/well for 1 h at 20 °C with shaking. Plates were washed eight times with wash buffer followed by the addition of enhancement solution (PerkinElmer, catalogue no. 1244-105). Plates were incubated for 5 min at 20 °C with shaking and the fluorescence, measured as cps (counts per second), was determined in a time-resolved fluorimeter (Victor™; PerkinElmer).
Western-blot analysis of ovine brain material
Ovine cerebellum was prepared in Check homogenization buffer (Prionics AG) prior to treatment with PK at a final concentration ranging from 0 to 32 μg/ml for 30 min at 37 °C. Digestion was terminated with 100 mM PMSF (Sigma, catalogue no. P-7626) and samples were subjected to SDS/PAGE run under reducing conditions (600 μg wet tissue equivalent per track) and subsequently transferred on to nitrocellulose membranes by semi-dry blotting. Membranes were blocked overnight at 4 °C with TBS-T (10 mM Tris/HCl, pH 7.8, 100 mM NaCl and 0.05% Tween 20) containing 5% (w/v) non-fat milk and subsequently washed three times with TBS-T. Membranes were incubated with purified anti-PrP mAb 683 (5 μg/ml) for 2 h at 20 °C and then washed five times with TBS-T. This was followed by incubation with goat anti-mouse IgG–HRP (horseradish peroxidase) (Sigma, catalogue no. A-3673) at 1:2000 for 2 h at 20 °C and five washes with TBS-T. All the antibody dilutions were prepared in 1% non-fat milk in TBS-T and the duration of each wash step was 5 min. PrP bands were detected by enhanced chemiluminescence (ECL®; Amersham Biosciences).
Nomenclature
Amino acid residue numbers refer to the ovine PrP sequence unless otherwise stated.
RESULTS
Development of a CDI
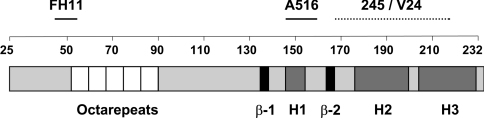
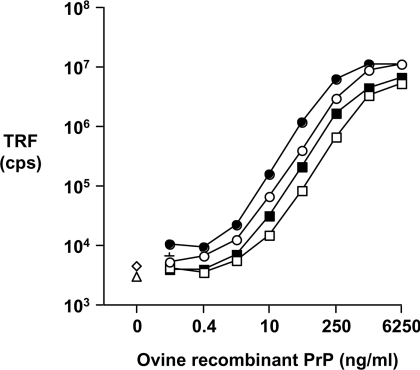
We have used a panel of anti-PrP mAbs to develop a novel CDI to characterize ovine PrPSc in brain samples from scrapie-infected sheep. N-terminal- and C-terminal-specific anti-PrP mAbs were tested in combination by capture-detector TRF-based immunoassay in order to establish pairs of reagents capable of efficient recognition of ovine PrP. Figure 1 shows the predicted epitope location of the N- and C-terminal-specific anti-PrP mAbs used in the present study. The N-terminal-specific capture mAb FH11 and the C-terminal-specific capture mAb A516 reacted well in combination with biotinylated C-terminal-specific mAb 245, V24 or V26 as detector reagent. These mAb pairs efficiently recognized both ARQ and VRQ allelic forms of ovine recombinant PrP protein. A representative standard curve for FH11 or A516 as capture anti-PrP mAbs and anti-PrP mAb 245 as detector is shown in Figure 2. The dose–response curves of the anti-PrP mAb pair FH11 and 245 were found to lie consistently to the left of those obtained for the mAb pair A516 and 245, which indicated an increased sensitivity of mAb pair FH11 and 245 for PrP. This may reflect the fact that FH11 binds within the N-terminal region of PrP, which is considered to be more solvent-exposed than the C-terminal region in which the epitope for A516 is located. Similar results were seen when the C-terminal-specific mAb V24 or V26 was used as the capture reagent (results not shown). Ovine recombinant ARQ protein was routinely recognized more efficiently than recombinant VRQ protein. Ovine recombinant VRQ protein appears to be a more compact molecule than the ARQ genotypic form and as a consequence specific epitopes are likely to be less accessible to antibody binding, in particular those in the C-terminal region of the protein. The overall sensitivity of the capture-detector immunoassays was in the region of 50 pg/ml of ovine recombinant PrP.
Figure 1. Epitope location of N- and C-terminal-specific anti-PrP mAbs.
β represents a β-strand and H represents a helix. Numbers represent amino acid residues 25–232.
Figure 2. Detection of ovine recombinant PrP by time-resolved capture-detector immunoassay.
Various concentrations of full-length ovine recombinant ARQ (filled symbols) or VRQ (open symbols) PrP protein was captured by anti-PrP mAb FH11-coated (circles) or A516-coated (squares) ELISA plates as described in the the Materials and methods section. Recombinant PrP was detected by 0.25 μg/ml purified anti-PrP mAb 245. Results shown are TRF cps±S.D. for triplicate wells.
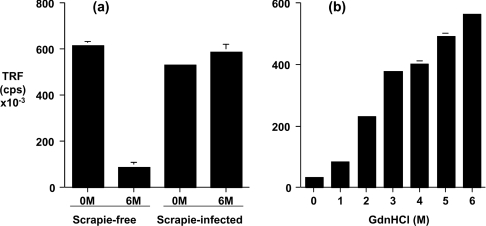
PrPC and PrPSc were detected by the capture-detector CDI following discrimination of the two prion protein isomers by denaturation-induced conformational changes in PrP epitope exposure. Figure 3(a) shows that significant levels of PrPC were detected in scrapie-free brain homogenate by the capture-detector immunoassay in the absence of any prior treatment of the tissue sample. This reflects the presence of N-terminal and C-terminal surface-exposed PrP epitopes in the normal isomer of PrP. This reactivity was lost when the scrapie-free brain homogenate was pretreated with 6 M GdnHCl. This reflects the loss of surface-exposed PrP epitopes within PrPC as a consequence of chaotropic-induced denaturation. In contrast, the level of fluorescence counts for scrapie-infected sheep brain homogenate treated with or without 6 M GdnHCl prior to the CDI was similar. The increase in fluorescence counts within the 6 M GdnHCl-treated scrapie-infected brain tissue sample compared with scrapie-free tissue reflects the exposure of previously buried epitopes within PrPSc, since either the FH11 or V24 epitope was no longer accessible in denatured PrPC.
Figure 3. Conformation-dependent immunodetection of ovine PrP.
(a) Scrapie-free and scrapie-infected homozygous VRQ brain homogenates were prepared as described in the Materials and methods section. Brain homogenates, treated with or without 6 M GdnHCl, were subjected to CDI using anti-PrP mAb FH11 as capture and anti-PrP mAb V24 as detector. (b) PrPSc was isolated from scrapie-infected homozygous VRQ brain homogenate by sarkosyl extraction and subjected to CDI following denaturation with increasing concentrations of GdnHCl. Results shown are TRF cps±S.D. for triplicate wells.
In order to confirm the selective recognition of ovine disease-associated PrP by denaturation-induced conformational changes, we performed the CDI on PrPSc isolated from scrapie-infected brain tissue. In order to do so, we made use of the fact that PrPC is soluble in detergent, whereas PrPSc is insoluble [32,33]. Accordingly, scrapie-infected brain homogenate was extracted with sarkosyl and aggregated, insoluble PrPSc was harvested by centrifugation at 21000 g. Native and denatured PrPSc were subjected to measurement by the CDI using anti-PrP mAbs FH11 and V24. Figure 3(b) shows that the immunoreactivity of sarkosyl-precipitated PrP was enhanced with increasing concentrations of GdnHCl. The maximum number of fluorescence counts was routinely achieved after treatment of PrPSc with 6 M GdnHCl. At this concentration of denaturant the CDI ratio (ratio of denatured/native cps) was also maximum. In combination with the absolute number of fluorescence counts measured, the CDI ratio provides a measure of total PrPSc and the relative level of PrPSc and PrPC within different preparations of tissue sample. Collectively, these observations show the utility of sarkosyl precipitation to isolate ovine PrPSc for analysis by CDI.
Measurement of ovine PrPSc by CDI
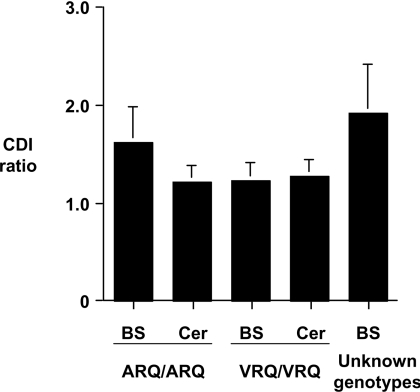
In order to investigate the level of aggregated PrP in brain tissue from sheep of different PrP genotypes, we first determined the CDI ratio of scrapie-negative samples. Brain stem and cerebellum were investigated to compare the CDI assay with standard diagnostic techniques utilizing only the brain stem. Results generated with cerebellar tissue would enable comparison with brain stem data to allow the potential to investigate various brain regions that may be targeted by different scrapie strains. Tissue from homozygous ARQ and VRQ scrapie-free sheep was extracted with sarkosyl, and precipitated material was subjected to the CDI using anti-PrP mAb FH11 as capture antibody and V24 as detector antibody. Low levels of fluorescence counts were obtained in both native and denatured samples of sarkosyl-precipitated PrP from brain stem and cerebellum of homozygous ARQ and VRQ New-Zealand-derived scrapie-free sheep, and from 44 unknown genotype abattoir brain stem samples that had all tested negative by the commercial Bio-Rad TeSeE test (results not shown). Figure 4 shows the CDI ratios for these scrapie-negative ovine brain samples. The mean CDI ratios for the scrapie-free homozygous ARQ and VRQ New-Zealand-derived samples were in the range 1.22–1.62 (Figure 4). Similar fluorescence counts and CDI values were obtained when anti-PrP mAb FH11 or A516 was used as capture in combination with either 245 or V24 as detector (results not shown). Collectively, these results indicate that little, if any, PrPC or aggregated PrP was precipitated from scrapie-free ovine brain material following sarkosyl extraction.
Figure 4. Low CDI values for scrapie-negative ovine brain tissue.
Scrapie-free brain homogenates were prepared from brain stem (BS) and cerebellum (Cer) of homozygous ARQ and VRQ sheep, and from brain stem of Bio-Rad TeSeE-negative sheep of unknown PrP genotypes, as described in the Materials and methods section. Brain homogenates, treated with or without 6 M GdnHCl, were subjected to CDI using mAb FH11 as capture and mAb V24 as detector. The results shown are CDI ratios (denatured/native fluorescence cps)±S.D. for scrapie-free brain stem and cerebellum brain tissue.
We subsequently examined the level of PrPSc in scrapie-infected brain stem and cerebellum from homozygous ARQ and VRQ scrapie-infected sheep by CDI. Figure 5 shows that significant levels of PrPSc were precipitated by sarkosyl treatment from both brain stem and cerebellum scrapie-infected homozygous VRQ sheep brain samples when measured by CDI using anti-PrP mAb FH11 as capture and V24 as detector. This was indicated by the increase in fluorescence counts for these different samples when treated with 6 M GdnHCl compared with that obtained in the absence of denaturant. In contrast, the level of fluorescence counts for both denatured and native samples obtained from scrapie-free brain samples was similar to background level. Similar fluorescence counts and CDI values were obtained when anti-PrP mAb FH11 or A516 was used as capture in combination with either 245 or V24 as detector (results not shown). These results indicate that significant levels of aggregated PrP are precipitated from scrapie-infected ovine brains following sarkosyl extraction.
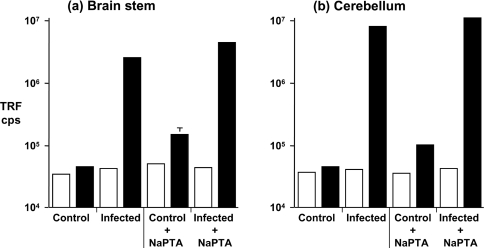
Figure 5. Detection of ovine PrPSc in (a) brain stem and (b) cerebellum by CDI.
Scrapie-free (control) and scrapie-infected (infected) homozygous VRQ brain stem and cerebellum homogenates were prepared as described in the Materials and methods section. PrPSc was isolated by sarkosyl extraction, with or without the use of sodium phosphotungstic acid (NaPTA) precipitation, and subjected to CDI using anti-PrP mAb FH11 as capture and anti-PrP mAb V24 as detector. Open bars: 0 M GdnHCl-treated samples; filled bars: 6 M GdnHCl-treated samples. Results shown are TRF cps±S.D. for triplicate wells.
In order to increase the sensitivity of the CDI we incorporated sodium phosphotungstic acid precipitation as a means to further enhance precipitation of disease-associated PrP. Sodium phosphotungstic acid precipitation has been reported to selectively precipitate PrPSc from a mixture of normal and abnormal isomers of PrP [17,34]. Figure 5 shows that when sodium phosphotungstic acid precipitation was used in conjunction with sarkosyl precipitation there was a significant increase in fluorescence counts within the 6 M GdnHCl-treated PrPSc isolated from both brain stem (Figure 5a) and cerebellum (Figure 5b). However, sodium phosphotungstic acid precipitation also increased the fluorescence counts in the 0 M fraction of scrapie-negative brain material, suggesting that this reagent does not selectively bind to PrPSc and may precipitate PrPC.
In order to address this further, homozygous ARR, ARQ and VRQ scrapie-free brain material was subjected to sarkosyl extraction, followed by sodium phosphotungstic acid precipitation, and the resultant PrP pellet was quantified by CDI. It was found that there was an increased CDI ratio for all three genotypes of ovine PrP brain material when sodium phosphotungstic acid precipitation was included as a procedure to extract PrP (results not shown). Interestingly, the highest CDI ratio was seen with homozygous VRQ brain material when sodium phosphotungstic acid precipitation was incorporated in the isolation of PrP, whereas in its absence the CDI ratios were similar between the three PrP genotypes. Since sodium phosphotungstic acid is believed to selectively precipitate abnormal isomers of PrP, this may indicate that VRQ PrP present in homozygous VRQ scrapie-free brain material is more likely to adopt such conformers. Because of these observations the use of sodium phosphotungstic acid was not routinely included in the isolation of PrPSc from non-PK-digested scrapie-infected brain samples.
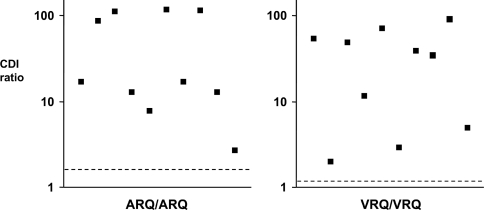
Having established a robust and reproducible assay to measure ovine disease-associated PrP in the absence of PK digestion, the level of PrPSc in natural cases of ovine scrapie was subsequently quantified by CDI. Homozygous ARQ and homozygous VRQ brain stem and cerebellum samples from sheep that were confirmed scrapie-positive cases were subjected to sarkosyl extraction and precipitated PrPSc was assessed by CDI using anti-PrP mAb FH11 as capture and V24 as detector. Isolated PrPSc was quantified in its native and denatured configuration and the CDI ratios for each sample were calculated. Figure 6 shows the CDI ratios for scrapie-infected brain stem material from homozygous ARQ (left-hand panel) and homozygous VRQ (right-hand panel) sheep. All of the known scrapie-positive samples of both genotypes showed a CDI ratio that was greater than the average of the genotype-equivalent scrapie-negative tissue. The range of CDI ratios for the samples from known scrapie-positive sheep were similar between the two genotypes and both sets of samples approximated into two clusters: those of low CDI ratios (in the range 3–30) and those with high CDI ratios (>30). The wide range of CDI ratios may reflect the variable distribution of PrPSc throughout the brain stem region as previously reported by Safar et al. [35]. It was therefore considered that the samples with high CDI ratios represented those that more accurately reflected the true level of PrPSc within scrapie-infected brain tissue. Accordingly, using standard curves of genotype-specific ovine recombinant PrP, the fluorescence counts obtained for denatured PrPSc for those samples with high CDI ratios (those >30) were expressed as nanogram equivalents of ovine recombinant PrP/mg of wet weight brain tissue. By this method, the amount of PrPSc in homozygous ARQ brain stem and cerebellum samples was calculated to be 120±20 and 108±67 ng/mg wet weight of tissue (n≥4) respectively, whereas for homozygous VRQ brain stem and cerebellum samples the levels were 450±250 and 517±272 ng/mg wet weight of tissue (n≥6) respectively.
Figure 6. CDI ratios for homozygous ARQ and VRQ scrapie-infected brain stems from natural cases of typical scrapie.
Homogenates were prepared from brain stem of known cases of homozygous ARQ (left-hand panel) and VRQ (right-hand panel) scrapie-infected sheep as described in the Materials and methods section. PrPSc was isolated by sarkosyl extraction and subjected to CDI using anti-PrP mAb FH11 as capture and anti-PrP mAb V24 as detector. The results shown are individual CDI ratios (denatured/native fluorescence cps) for each tissue sample. The broken line is the mean CDI value for scrapie-free tissue of the same genotype (n=10 for each genotype).
Detection of PK-sensitive PrPSc in scrapie-infected brain material
We have isolated PrPSc without the use of PK for quantification by CDI. This allowed us to investigate whether ovine PrPSc comprised PK-sensitive and -resistant PrPSc material and if the relative levels of these different conformers of PrPSc showed genotypic variation in scrapie-infected ovine brain tissue. In order to do so, we first analysed the susceptibility of ovine PrP to PK digestion.
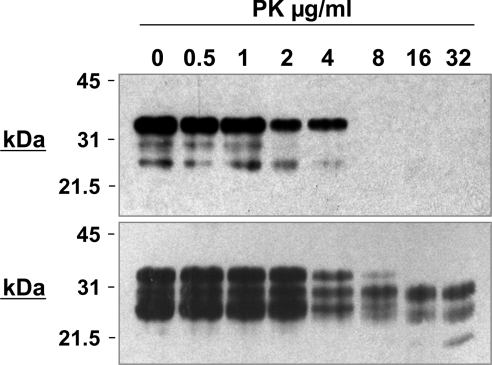
Figure 7 shows the effect of various concentrations of PK enzyme on scrapie-free (upper panel) and scrapie-infected (lower panel) homozygous VRQ brain material. All of the PrPC within the scrapie-free brain material was digested by concentrations of PK equal to, or greater than, 8 μg/ml. When scrapie-infected brain material was digested with similar concentrations of the proteolytic enzyme, PK-resistant PrPSc was evident at concen-trations of PK equal to, or greater than, 16 μg/ml. Although not formally quantified, it was clear that the PK-resistant material constituted a minor component of the brain sample analysed. Similar results were seen when homozygous ARQ scrapie-free and scrapie-infected brain material was analysed in a similar manner (results not shown).
Figure 7. Detection of ovine PrP by Western blot.
Homozygous VRQ scrapie-free (upper panel) and scrapie-infected (lower panel) brain material was prepared as described in the Materials and methods section. Brain homogenate was treated with various concentrations of PK and analysed by SDS/PAGE and Western blot using anti-PrP mAb 683; 600 μg wet tissue equivalent was loaded per track. Molecular mass markers (kDa) are shown on the left-hand side.
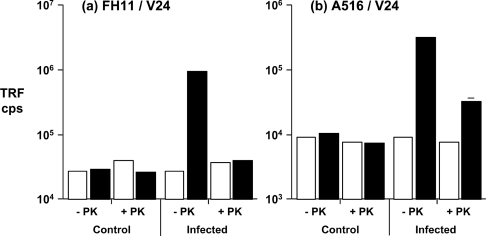
Accordingly, scrapie-infected ovine brain samples were subjected to sarkosyl extraction and precipitated PrPSc was quantified by CDI, before and after treatment of the samples with PK. Figure 8 shows that non-PK-digested precipitated PrPSc was efficiently recognized when the N-terminal-specific mAb FH11 was used in combination with the C-terminal-specific mAb V24 for CDI (Figure 8a). However, the immunoreactivity with this mAb pair was lost following treatment of PrPSc with PK. When the C-terminal mAb A516 was used for capture, reactivity with disease-associated PrP was seen before and after digestion of the sample with PK. However, the level of PrPSc detected by CDI using anti-PrP mAb pair A516 and V24 following PK digestion was approx. 10% of that seen in the absence of protease digestion (Figure 8b). These observations indicated that most of the sarkosyl-precipitated ovine PrPSc was PK sensitive. The results also show that a minor fraction of PrPSc was an N-terminally truncated C-terminal protease-resistant fragment, consistent with material designated PrP27–30 that correlates with the PK-resistant PrPSc shown by Western blot (Figure 7).
Figure 8. Detection of PK-sensitive ovine PrPSc by CDI.
Scrapie-free (control) and scrapie-infected (infected) homozygous VRQ brain stem homogenates were prepared as described in the Materials and methods section. PrPSc was isolated by sarkosyl extraction, treated with (+) or without (−) 16 μg/ml of PK, and subjected to CDI using (a) anti-PrP mAb FH11 or (b) anti-PrP mAb A516 as capture, and in both cases anti-PrP mAb V24 as detector. Open bars: 0 M GdnHCl-treated samples; filled bars: 6 M GdnHCl-treated samples. Results shown are TRF cps±S.D. for triplicate wells.
Having established the presence of PK-sensitive and PK-resistant PrPSc in brain material from scrapie-infected sheep, we subsequently investigated whether there were any differences in the level of these different conformers in natural scrapie samples from sheep of different genotypes. PrPSc isolated from homozygous ARQ and VRQ scrapie-infected brain samples by sarkosyl precipitation was treated with or without PK digestion followed by CDI measurement, as shown in Figure 8. The level of PK-resistant PrPSc in homozygous ARQ scrapie-infected brain samples was 56±34% for brain stem and 41±12% for cerebellum (n=4 in both cases). The level of PK-resistant PrPSc in homozygous VRQ scrapie-infected brain samples was 35±28% for brain stem and 27±27% for cerebellum (n=4 in both cases). The remainder of the PrPSc detected was PK-sensitive material. Therefore, by subtraction, the mean levels of PK-sensitive material in the brain stem of homozygous ARQ and VRQ animals were approx. 44 and 65% respectively. Similarly, in the cerebellum, the mean levels of PK-sensitive material were approx. 59 and 73% for homozygous ARQ and VRQ animals respectively. The standard deviations within these data were high, in some cases as wide as the absolute reading of PK-resistant PrPSc, presumably due to a high level of animal-to-animal variation in the PrPSc fractions measured. Despite this variation in the level of PrPSc detected within samples from the same genotype, these data do indicate a trend for higher levels of PK-sensitive PrPSc in homozygous VRQ naturally scrapie-infected sheep brains. However, there would appear to be higher absolute levels of both PK-sensitive and PK-resistant PrPSc conformers in homozygous VRQ scrapie-infected ovine brain material since the level of total PrPSc in these animals was found to be approximately twice that in homozygous ARQ sheep.
DISCUSSION
A feature of scrapie disease is that phenotypically distinct prion strains can be identified that show unique biological properties. According to the protein-only hypothesis, strain variation is attributed to the existence of conformational subtypes of disease-associated PrP [17,36–38]. Accordingly, biochemical and immunological approaches have been developed to aid the distinction of prion subtypes through the molecular analysis of PrPSc. These approaches include assessment of relative resistance to PK, molecular mass of the PK-resistant core, thermodynamic stability and epitope accessibility [16–18,36,37,39]. Conformations of PrP have been studied through the use of anti-PrP mAbs to analyse variation in epitope presentation during denaturation or limited PK digestion of the prion protein. Here, we have developed a CDI to study the various conformers of different genotypes of ovine PrPSc as a prelude to biochemical and molecular strain typing of ovine scrapie. We have tested tissues routinely used for scrapie diagnosis that are expected to harbour reasonable levels of PrPSc found in sheep infected with either typical or atypical scrapie strains. The CDI developed here has utilized two features of PrPSc: first, its susceptibility to be precipitated from brain homogenates by detergent, whereas PrPC is solubilized; secondly, that surface-exposed epitopes are buried in native or aggregated PrPSc but are made accessible following denaturation of the molecule.
Detergent extraction has proved to be a simple and effective procedure for the isolation of PrPSc from ovine scrapie-infected brain tissue. The procedure was selective for disease-associated PrP since very little, if any, aggregated detergent-insoluble PrP was precipitated from scrapie-free ovine brain material. Using this technique coupled with immunodetection by CDI, we have shown here that brain material from homozygous VRQ sheep with terminal natural scrapie was characterized by higher levels of PrPSc than their counterpart homozygous ARQ samples. A similar phenomenon has been reported for these genotypes of sheep infected with experimental scrapie [40]. The occurrence of higher levels of PrPSc in homozygous VRQ sheep occurs despite the fact that these animals succumb to natural and experimental scrapie with a shorter incubation time compared with homozygous ARQ animals. Several possibilities exist for the increased level of PrPSc in homozygous VRQ scrapie-infected brains. First, increased CNS expression of PrPC protein in homozygous VRQ sheep could provide more substrate for the conversion of normal PrP into PrPSc. However, our own Western-blot analysis of scrapie-free sheep brain samples has shown similar levels of PrPC in homozygous VRQ sheep brain samples compared with homozygous ARQ and ARR genotypes [41]. Secondly, VRQ PrPC could be more resistant to metabolic breakdown compared with ARQ PrPC and consequently may be associated with a longer cellular half-life. This is supported by our own observations and those of others that have shown that ovine VRQ PrP is more compact than A136 ovine PrP [24–26]. The fact that the VRQ allelic variant has a high propensity for β-sheet formation [42] could consequently render the protein more susceptible to accumulate in the cell in a disease-associated form. Thirdly, the increased stability of the VRQ allelic variant may also be evident in the PrPSc form of this protein. If this is the case, higher levels of VRQ PrPSc may accumulate as a consequence of a lower rate of clearance compared with other allelic forms of ovine PrPSc. These possibilities are not mutually exclusive and concur with the view that differences in the metabolism of allelic variants of ovine PrP contribute to the mechanism(s) that determine susceptibility and resistance of sheep to natural scrapie [24,25] and that the levels of brain PrPSc are determined by both the rates of formation and clearance [43].
PrPSc is typically distinguished in a mixture with PrPC as a relative-protease-resistant moiety following proteolysis of the normal PrP isomer with PK. Here, we excluded the use of PK in order to identify those conformations of ovine disease-associated PrP in natural cases of scrapie that may be more sensitive to proteolytic digestion than other forms. Our results show that ovine PrPSc from natural cases of typical scrapie consisted of both PK-sensitive and PK-resistant material. PrPSc comprised up to 90% PK-sensitive material in some cases. The relative contribution of PK-sensitive and PK-resistant PrPSc conformers appeared to differ between scrapie-infected sheep of different genotypes, since homozygous VRQ samples appeared to be characterized by a higher percentage of PK-sensitive material than homozygous ARQ samples. This would be consistent with the view that V136 PrP has a longer cellular half-life than A136 allelic forms of ovine PrP [24–26,41]. There has been considerable debate in recent years on whether mature amyloid fibrils and/or soluble oligomers of PrPSc represent the infectious prion agent and the molecule responsible for neurotoxicity seen during the progression of prion disease [7,44,45]. A prevailing view is that the deposition of mature fibrillar amyloid aggregates of PrPSc is a protective mechanism that exists to avoid the high intrinsic toxicity of soluble oligomers [44,46,47]. According to such a scheme, the more rapid progression of typical scrapie disease in homozygous VRQ sheep should correlate with the presence of soluble infectious and toxic PrP oligomers in the PK-sensitive fraction of ovine PrPSc. This remains to be determined.
The necessity to detect PK-sensitive forms of PrPSc has been highlighted through the emergence of atypical scrapie in PrP genotypes of sheep that have shown resistance to classical or typical scrapie, including homozygous ARR sheep [48–51]. A distinguishing feature of atypical sheep scrapie is that the associated PrPSc shows a significantly lower level of resistance to digestion with PK than does PrPSc from typical sheep scrapie cases [52]. The relative PK-sensitive atypical PrPSc does transmit to ovine PrP transgenic mice, indicating that this material is indeed infectious [52]. This suggests that the use of PK to identify disease-associated PrPSc may only uncover a limited part of the spectrum of infectious prion conformers in scrapie disease of sheep. In support of this view, other forms of TSE disease have shown variation in PK resistance between strains, such as ‘drowsy’ and ‘hyper’ in prions in transmissible mink encephalopathy [53]. Accordingly, distinct prion strains of sheep scrapie, which according to the protein-only hypothesis are synonymous with distinct conformers of PrPSc, may exist that are characterized by an even greater sensitivity to proteolytic digestion than is PrPSc from the presently identified atypical sheep scrapie cases. These prion strains will clearly not be identified by those current rapid tests that all utilize digestion with PK to identify disease-associated PrP in scrapie-infected sheep.
We have investigated the use of sodium phosphotungstic acid precipitation in an attempt to increase the yield of PrPSc extracted from scrapie-infected sheep brain tissue for measurement by CDI. Sodium phosphotungstic acid precipitation did increase the level of fluorescence counts in the 6 M GdnHCl fraction of sarkosyl-precipitated scrapie-infected brain homogenates but this was also seen with scrapie-free brain material. In addition, sodium phosphotungstic acid precipitation also increased fluorescence counts of the 0 M GdnHCl fraction of sarkosyl-extracted scrapie-free sheep brain homogenates. Collectively, these results suggest that under the conditions used here, sodium phosphotungstic acid precipitated PrPC as well as PrPSc from sheep brain homogenates. This may have been due to the presence of Mg2+ ions, which have been reported to reduce the selectivity of PrPSc isolation [54]. However, despite this, higher levels of PrP were precipitated by the use of sodium phosphotungstic acid from homozygous VRQ scrapie-free brain tissue compared with homozygous ARQ and ARR brain samples. This difference could reflect the presence of distinct PrP conformers in homozygous VRQ scrapie-free brains compared with those in brain tissue from A136 genotypes. It has been shown that low concentrations of sarkosyl, and other anionic detergents, are capable of inducing aggregation and increased β-sheet content of normal PrPC, without the acquisition of PK-resistance or infectious potential [55,56]. However, the concentrations of sarkosyl used to induce aggregation and β-sheet formation in PrPC were significantly less than those used to extract PrPSc from ovine brain tissue [33,56]. It is envisaged that at low concentrations of sarkosyl there is a partial denaturation of PrPC, while at higher concentrations there is more complete disruption of the hydrophobic core accompanied by burial of the hydrophobic surface in the detergent micelles, aiding solubilization of the protein. The fact that some PrP was precipitated from scrapie-free sheep brains may suggest that aggregates of normal PrPC do exist in normal brain tissue. This material did not appear to be protease-resistant as there was no difference in fluorescence counts before and after its digestion with PK when detected by the C-terminal anti-PrP mAb pair A516 and V24. However, the fact that more material was precipitated from homozygous VRQ brain material compared with A136 allelic variant tissue correlates with our observation that ovine recombinant VRQ PrP has a greater potential to form β-sheet structure [42].
The development of a CDI that discriminates different conformers of ovine PrPSc allows the quantification of different PrPSc subtypes and their correlation with different properties of the protein such as prion infectivity. This is a highly relevant issue since it has been shown in other systems that prion infectivity and the level of PK-resistant PrPSc do not show absolute correlation [12–15]. Furthermore, the incubation period of different strains of hamster prions were found to correlate with the level of PK-sensitive rather than PK-resistant PrPSc. Whether ovine prion strains can be defined in a similar manner by their level of PK-resistant and PK-sensitive PrPSc remains to be established. In our attempts to address this, we have measured the level of PrPSc in homozygous ARQ and VRQ scrapie-infected brain homogenates that have previously been designated ‘good’ or ‘poor’ transmitters following inoculation into wild-type mice. Quantification of PrPSc by CDI has shown that both ‘good’ and ‘poor’ transmitters contained similar levels of total PrPSc, whereas only the ‘good’ transmitters contained significant levels of PK-resistant PrPSc as indicated by Western blot (A. M. Thackray, L. Hopkins and R. Bujdoso, unpublished work). Collectively, these results suggest that the incubation time for scrapie correlates with the level of distinct conformations of PrPSc. This type of analysis provides a basis to investigate the role of different PrP conformations in the biochemical description of different ovine scrapie strains.
Acknowledgments
This work was supported by funds from Defra (Department for Environment, Food and Rural Affairs). L. H. is in receipt of a Defra Ph.D. studentship. We thank Dr C. R. Birkett (Institute for Animal Health, Compton Laboratories, Newbury, U.K.) and the TSE Resource Centre for mAb FH11. We thank the TSE Archive, Veterinary Laboratories Agency Weybridge (Addlestone, Surrey, U.K.), for the supply of scrapie-free and scrapie-infected tissue samples, and ADAS (Shrewsbury, U.K.), for scrapie-negative abattoir samples.
References
- 1.Prusiner S. B. Early evidence that a protease-resistant protein is an active component of the infectious prion. Cell. 2004;116:S109. doi: 10.1016/s0092-8674(03)01032-8. [DOI] [PubMed] [Google Scholar]
- 2.Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S. B., McKinley M. P., Bowman K. A., Bolton D. C., Bendheim P. E., Groth D. F., Glenner G. G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 4.Caughey B. W., Dong A., Bhat K. S., Ernst D., Hayes S. F., Caughey W. S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 5.Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 7.Legname G., Baskakov I. V., Nguyen H. O., Riesner D., Cohen F. E., DeArmond S. J., Prusiner S. B. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 8.Castilla J., Saa P., Hetz C., Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner S. B. Prion Biology and Diseases. 2nd edn. Plainview, NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 10.McKinley M. P., Bolton D. C., Prusiner S. B. A protease-resistant protein is a structural component of the scrapie prion. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 11.Hope J., Morton L. J., Farquhar C. F., Multhaup G., Beyreuther K., Kimberlin R. H. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP) EMBO J. 1986;5:2591–2597. doi: 10.1002/j.1460-2075.1986.tb04539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barron R. M., Thomson V., Jamieson E., Melton D. W., Ironside J., Will R., Manson J. C. Changing a single amino acid in the N-terminus of murine PrP alters TSE incubation time across three species barriers. EMBO J. 2001;20:5070–5078. doi: 10.1093/emboj/20.18.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lasmezas C. I., Deslys J. P., Robain O., Jaegly A., Beringue V., Peyrin J. M., Fournier J. G., Hauw J. J., Rossier J., Dormont D. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 14.Manuelidis L., Fritch W., Xi Y. G. Evolution of a strain of CJD that induces BSE-like plaques. Science. 1997;277:94–98. doi: 10.1126/science.277.5322.94. [DOI] [PubMed] [Google Scholar]
- 15.Tremblay P., Ball H. L., Kaneko K., Groth D., Hegde R. S., Cohen F. E., DeArmond S. J., Prusiner S. B., Safar J. G. Mutant PrPSc conformers induced by a synthetic peptide and several prion strains. J. Virol. 2004;78:2088–2099. doi: 10.1128/JVI.78.4.2088-2099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., et al. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J. Mol. Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 17.Safar J., Wille H., Itri V., Groth D., Serban H., Torchia M., Cohen F. E., Prusiner S. B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 18.Paramithiotis E., Pinard M., Lawton T., LaBoissiere S., Leathers V. L., Zou W. Q., Estey L. A., Lamontagne J., Lehto M. T., Kondejewski L. H., et al. A prion protein epitope selective for the pathologically misfolded conformation. Nat. Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 19.Baylis M., Chihota C., Stevenson E., Goldmann W., Smith A., Sivam K., Tongue S., Gravenor M. B. Risk of scrapie in British sheep of different prion protein genotype. J. Gen. Virol. 2004;85:2735–2740. doi: 10.1099/vir.0.79876-0. [DOI] [PubMed] [Google Scholar]
- 20.Belt P. B., Muileman I. H., Schreuder B. E., Bos-de Ruijter J., Gielkens A. L., Smits M. A. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J. Gen. Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 21.Goldmann W., Hunter N., Smith G., Foster J., Hope J. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J. Gen. Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda T., Horiuchi M., Ishiguro N., Muramatsu Y., Kai-Uwe G. D., Shinagawa M. Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J. Gen. Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 23.Bossers A., Belt P., Raymond G. J., Caughey B., de Vries R., Smits M. A. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rezaei H., Choiset Y., Eghiaian F., Treguer E., Mentre P., Debey P., Grosclaude J., Haertle T. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 2002;322:799–814. doi: 10.1016/s0022-2836(02)00856-2. [DOI] [PubMed] [Google Scholar]
- 25.Eghiaian F., Grosclaude J., Lesceu S., Debey P., Doublet B., Treguer E., Rezaei H., Knossow M. Insight into the PrPC→PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10254–10259. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bujdoso R., Burke D. F., Thackray A. M. Structural differences between allelic variants of the ovine prion protein revealed by molecular dynamics simulations. Proteins. 2005;61:840–849. doi: 10.1002/prot.20755. [DOI] [PubMed] [Google Scholar]
- 27.Benestad S. L., Sarradin P., Thu B., Schonheit J., Tranulis M. A., Bratberg B. Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet. Rec. 2003;153:202–208. doi: 10.1136/vr.153.7.202. [DOI] [PubMed] [Google Scholar]
- 28.Thackray A. M., Yang S., Wong E., Fitzmaurice T. J., Morgan-Warren R. J., Bujdoso R. Conformational variation between allelic variants of cell-surface ovine prion protein. Biochem. J. 2004;381:221–229. doi: 10.1042/BJ20040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster J. D., Wilson M., Hunter N. Immunolocalisation of the prion protein (PrP) in the brains of sheep with scrapie. Vet. Rec. 1996;139:512–515. doi: 10.1136/vr.139.21.512. [DOI] [PubMed] [Google Scholar]
- 30.Thuring C. M., van Keulen L. J., Langeveld J. P., Vromans M. E., van Zijderveld F. G., Sweeney T. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J. Comp. Pathol. 2005;132:59–69. doi: 10.1016/j.jcpa.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Thackray A. M., Madec J. Y., Wong E., Morgan-Warren R., Brown D. R., Baron T., Bujdoso R. Detection of bovine spongiform encephalopathy, ovine scrapie prion-related protein (PrPSc) and normal PrPc by monoclonal antibodies raised to copper-refolded prion protein. Biochem. J. 2003;370:81–90. doi: 10.1042/BJ20021280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilmert H., Diringer H. A rapid and efficient method to enrich SAF-protein from scrapie brains of hamsters. Biosci. Rep. 1984;4:165–170. doi: 10.1007/BF01120313. [DOI] [PubMed] [Google Scholar]
- 33.McKinley M. P., Meyer R. K., Kenaga L., Rahbar F., Cotter R., Serban A., Prusiner S. B. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J. Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wadsworth J. D., Joiner S., Hill A. F., Campbell T. A., Desbruslais M., Luthert P. J., Collinge J. Tissue distribution of protease resistant prion protein in variant Creutzfeldt–Jakob disease using a highly sensitive immunoblotting assay. Lancet. 2001;358:171–180. doi: 10.1016/s0140-6736(01)05403-4. [DOI] [PubMed] [Google Scholar]
- 35.Safar J. G., Scott M., Monaghan J., Deering C., Didorenko S., Vergara J., Ball H., Legname G., Leclerc E., Solforosi L., et al. Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat. Biotechnol. 2002;20:1147–1150. doi: 10.1038/nbt748. [DOI] [PubMed] [Google Scholar]
- 36.Bessen R. A., Marsh R. F. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peretz D., Scott M. R., Groth D., Williamson R. A., Burton D. R., Cohen F. E., Prusiner S. B. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telling G. C., Parchi P., DeArmond S. J., Cortelli P., Montagna P., Gabizon R., Mastrianni J., Lugaresi E., Gambetti P., Prusiner S. B. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 39.Bellon A., Seyfert-Brandt W., Lang W., Baron H., Groner A., Vey M. Improved conformation-dependent immunoassay: suitability for human prion detection with enhanced sensitivity. J. Gen. Virol. 2003;84:1921–1925. doi: 10.1099/vir.0.18996-0. [DOI] [PubMed] [Google Scholar]
- 40.McCutcheon S., Hunter N., Houston F. Use of a new immunoassay to measure PrP Sc levels in scrapie-infected sheep brains reveals PrP genotype-specific differences. J. Immunol. Methods. 2005;298:119–128. doi: 10.1016/j.jim.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Thackray A. M., Fitzmaurice T. J., Hopkins L., Bujdoso R. Ovine plasma prion protein levels show genotypic variation detected by C-terminal epitopes not exposed in cell-surface PrPC. Biochem. J. 2006;400:349–358. doi: 10.1042/BJ20060746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong E., Thackray A. M., Bujdoso R. Copper induces increased beta-sheet content in the scrapie-susceptible ovine prion protein PrPVRQ compared with the resistant allelic variant PrPARR. Biochem. J. 2004;380:273–282. doi: 10.1042/BJ20031767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safar J. G., DeArmond S. J., Kociuba K., Deering C., Didorenko S., Bouzamondo-Bernstein E., Prusiner S. B., Tremblay P. Prion clearance in bigenic mice. J. Gen. Virol. 2005;86:2913–2923. doi: 10.1099/vir.0.80947-0. [DOI] [PubMed] [Google Scholar]
- 44.Anderson M., Bocharova O. V., Makarava N., Breydo L., Salnikov V. V., Baskakov I. V. Polymorphism and ultrastructural organization of prion protein amyloid fibrils: an insight from high resolution atomic force microscopy. J. Mol. Biol. 2006;358:580–596. doi: 10.1016/j.jmb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Legname G., Nguyen H. O., Baskakov I. V., Cohen F. E., Dearmond S. J., Prusiner S. B. Strain-specified characteristics of mouse synthetic prions. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan J. C., Oyler N. A., Yau W. M., Tycko R. Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luhrs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Dobeli H., Schubert D., Riek R. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buschmann A., Biacabe A. G., Ziegler U., Bencsik A., Madec J. Y., Erhardt G., Luhken G., Baron T., Groschup M. H. Atypical scrapie cases in Germany and France are identified by discrepant reaction patterns in BSE rapid tests. J. Virol. Methods. 2004;117:27–36. doi: 10.1016/j.jviromet.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 49.De Bosschere H., Roels S., Dechamps P., Vanopdenbosch E. TSE detected in a Belgian ARR-homozygous sheep via active surveillance. Vet. J. 2005. in the press. [DOI] [PubMed]
- 50.Everest S. J., Thorne L., Barnicle D. A., Edwards J. C., Elliott H., Jackman R., Hope J. Atypical prion protein in sheep brain collected during the British scrapie-surveillance programme. J. Gen. Virol. 2006;87:471–477. doi: 10.1099/vir.0.81539-0. [DOI] [PubMed] [Google Scholar]
- 51.Gavier-Widen D., Noremark M., Benestad S., Simmons M., Renstrom L., Bratberg B., Elvander M., af Segerstad C. H. Recognition of the Nor98 variant of scrapie in the Swedish sheep population. J. Vet. Diagn. Invest. 2004;16:562–567. doi: 10.1177/104063870401600611. [DOI] [PubMed] [Google Scholar]
- 52.Le Dur A., Beringue V., Andreoletti O., Reine F., Lai T. L., Baron T., Bratberg B., Vilotte J. L., Sarradin P., Benestad S. L., et al. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16031–16036. doi: 10.1073/pnas.0502296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bessen R. A., Marsh R. F. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee I. S., Long J. R., Prusiner S. B., Safar J. G. Selective precipitation of prions by polyoxometalate complexes. J. Am. Chem. Soc. 2005;127:13802–13803. doi: 10.1021/ja055219y. [DOI] [PubMed] [Google Scholar]
- 55.May B. C., Govaerts C., Prusiner S. B., Cohen F. E. Prions: so many fibers, so little infectivity. Trends Biochem. Sci. 2004;29:162–165. doi: 10.1016/j.tibs.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Xiong L. W., Raymond L. D., Hayes S. F., Raymond G. J., Caughey B. Conformational change, aggregation and fibril formation induced by detergent treatments of cellular prion protein. J. Neurochem. 2001;79:669–678. doi: 10.1046/j.1471-4159.2001.00606.x. [DOI] [PubMed] [Google Scholar]