Figure 1. Panel of soluble gammaretroviral envelope RBDs and SUs tested as entry cofactors.
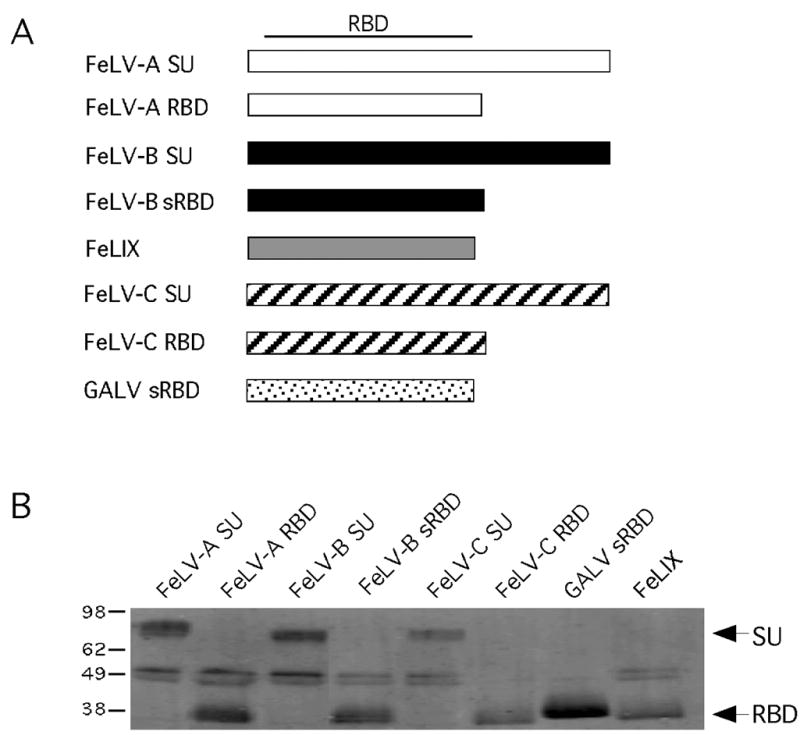
(A) Schematic representation of the structure of each envelope-derived cofactor. The name of each cofactor is indicated to the left of the boxes representing the linear DNA sequence. The approximate location of the receptor binding domain (RBD) within the SU is indicated at the top. (B) Western blot analysis of a volume of conditioned supernatant that corresponds to one equivalent unit of each envelope-derived cofactor. The amount loaded in each lane was determined from the results of quantitative Western blot analysis of equal amounts of each supernatant (not shown). Envelope-derived cofactors were detected with the monoclonal antibody HA.11 to the C-terminal HA-epitope tags as described in Materials and Methods. To the left of the blot are approximate kilodaltons; to the right are the expected sizes of the SU and sRBD constructs.
