Abstract
The Myc oncoprotein is a potent inducer of cell growth, cell cycle progression, and apoptosis. While many direct Myc target genes have been identified, the molecular determinants of Myc’s transcriptional specificity remain elusive. We have carried out a genetic screen in Drosophila and identified the Trithorax group protein Little imaginal discs (Lid) as a regulator of dMyc-induced cell growth. Lid binds to dMyc and is required for dMyc-induced expression of the growth regulatory gene Nop60B. The mammalian Lid orthologs, Rbp-2 (JARID1A) and Plu-1 (JARID1B), also bind to c-Myc, indicating that Lid–Myc function is conserved. We demonstrate that Lid is a JmjC-dependent trimethyl H3K4 demethylase in vivo and that this enzymatic activity is negatively regulated by dMyc, which binds to Lid’s JmjC domain. Because Myc binding is associated with high levels of trimethylated H3K4, we propose that the Lid–dMyc complex facilitates Myc binding to, or maintenance of, this chromatin context.
Keywords: Myc, little imaginal discs, Trithorax group (TrxG), JmjC, Drosophila, histone demethylase
The myc proto-oncogene family and its encoded proteins are thought to function as both sensors and mediators of many growth and mitogenic signals (Grandori et al. 2000; Luscher 2001; Oster et al. 2002). Multiple signal transduction pathways have been shown to influence myc gene transcription (Liu and Levens 2006), and some signaling pathways have been shown to regulate the levels of Myc protein through proteosome-mediated degradation (Kenney et al. 2003; Yeh et al. 2004). Thus the abundance of Myc protein serves as a read-out and integrator of a wide range of environmental signals. Myc protein, in turn, coordinates the cellular responses to these environmental signals by functioning as a transcription factor that modulates the expression of hundreds of genes (Patel et al. 2004; Adhikary and Eilers 2005). Indeed, numerous expression array studies have shown that Myc induces a relatively weak transcriptional response of a surprisingly large number of genes. Many of these genes appear to be direct targets of Myc protein as determined by genomic profiling assays in Drosophila (Orian et al. 2003) and mammalian cells (Fernandez et al. 2003; Li et al. 2003). Myc’s transcriptional targets include many genes involved in cell growth (ribosome biogenesis, translation, metabolism), a lesser number of cell cycle genes, and genes encoding clusters of microRNAs (Coller et al. 2000; Guo et al. 2000; Schuhmacher et al. 2001; O’Connell et al. 2003; He et al. 2005; O’Donnell et al. 2005). Moreover, Myc has been demonstrated to influence transcription by all three nuclear RNA polymerases (Oskarsson and Trumpp 2005). In both mammalian and Drosophila cells, increasing Myc levels stimulates rDNA transcription by RNA polymerase I as an integral feature of the augmented cell growth response to Myc (Arabi et al. 2005; Grandori et al. 2005; Grewal et al. 2005).
Given the diversity of its transcriptional targets, it is not surprising that Myc has been implicated in several distinct modes of interaction with chromatin and with the transcriptional apparatus. Myc belongs to a larger class of basic region helix–loop–helix–zipper (bHLHZ) proteins and has been demonstrated to form a highly specific heterodimer with the small bHLHZ protein Max. The Myc–Max dimer exhibits sequence-specific DNA binding to the E-box sequence CACGTG and lower-affinity binding to several other related sequences (for recent reviews, see Oster et al. 2002; Adhikary and Eilers 2005; Cole and Nikiforov 2006). In general, binding of Myc–Max to E-boxes within the vicinity of a promoter leads to transcriptional activation. Such activity requires the Myc bHLHZ domain (for Max interaction) and several highly conserved regions proximal to the N terminus of the Myc protein. One of these regions, Myc Box II (MBII), has been shown to associate with a large coactivator known as TRAPP (McMahon et al. 1998), which in turn binds the histone acetyltransferase GCN5 and other members of the SAGA (SPT/ADA/GCN5/Acetyltransferase) complex. Distinct TRAPP complexes containing the Tip60 acetyltransferase along with the ATPases Tip48 and Tip49 also associate with Myc (Wood et al. 2000; Frank et al. 2003). The association of these proteins with Myc and the demonstration that they are required for at least some Myc functions suggests that histone acetylation represents an important aspect of Myc’s effect on transcription (for reviews, see Amati et al. 2001; Cole and Nikiforov 2006). This idea has also received strong support from chromatin immunoprecipitation experiments demonstrating that Myc binding and acetylation of promoter-proximal histones is associated with transcriptional activation of many Myc target genes (Bouchard et al. 2001; Frank et al. 2001; Orian et al. 2005).
While TRRAP complexes are undoubtedly important for Myc transcriptional activity, there is also evidence that activation of some Myc target genes is independent of TRRAP binding (Nikiforov et al. 2002). Activation of the TERT and cad promoters by c-Myc has been reported to be largely due to stimulation of transcriptional elongation by P-TEFb (cyclin T1–cdk9 complex) leading to hyperphosphorylation of the RNA polymerase II C-terminal domain (Bouchard et al. 2001; Eberhardy and Farnham 2001, 2002). There is also circumstantial evidence that chromatin remodeling complexes may play a role in Myc function. The Tip48 and Tip49 ATPases that have been implicated in chromatin remodeling in yeast (Shen et al. 2000) have been found to interact with Myc, both as subunits of TRRAP complexes and independently of TRRAP, in mammalian and Drosophila cells (Wood et al. 2000; Bellosta et al. 2005). Furthermore, the SWI/SNF subunit INI1/hSNF5 has been reported to associate with the c-Myc bHLHZ region (Cheng et al. 1999). However, changes in nucleosome positioning have yet to be demonstrated to accompany Myc binding and activation.
Myc has also been shown to be involved in transcriptional repression. Interestingly, many of the genes thought to be down-regulated by Myc encode proteins involved in cell cycle arrest. These include a subset of cyclin-dependent kinase inhibitors (e.g., p21CIP1, p15INK4B), differentiation inducers, and proteins linked to growth arrest in response to stress (for review, see Kleine-Kohlbrecher et al. 2006). While several possible molecular mechanisms for Myc repression have been suggested, the most compelling and well documented involves direct interaction of Myc–Max heterodimers with the BTB-POZ domain protein Miz-1. The interaction serves to inhibit the transcriptional activity of Miz-1 at its target gene promoters (such as p21CIP1 and p15INK4b), resulting in repression. The Miz-1 association with Myc has been shown to be important in modulating the growth inhibitory responses to TGFb and to UV damage as well as in keratinocyte differentiation (Seoane et al. 2001; Staller et al. 2001; Herold et al. 2002; Gebhardt et al. 2006).
While the studies summarized above indicate that Myc participates in several distinct transcription-related complexes, it remains uncertain whether these are the only, or the most critical, complexes involved in Myc function. In particular, the recent findings that Myc binds on the order of 15% of genomic loci and has broad effects on chromatin structure (Fernandez et al. 2003; Li et al. 2003; Orian et al. 2003, 2005; Knoepfler et al. 2006) raise the possibility that Myc may have an impact on other chromatin-regulatory proteins to influence the spreading of histone modifications over larger chromatin domains. Over the last decade, we and others have exploited Drosophila melanogaster to study the activity and functions of dMyc, the single Drosophila ortholog of vertebrate Myc (for review, see Gallant 2006). dMyc, like vertebrate Myc proteins, dimerizes with dMax and binds E-box sequences to activate transcription. It also binds widely throughout the fly genome and activates a broad program of gene expression related to cell growth (Johnston et al. 1999; Orian et al. 2005). Furthermore, dMyc and c-Myc have been shown to be functionally interchangeable in assays of transformation, proliferation, and apoptosis (Schreiber-Agus et al. 1997; Trumpp et al. 2001; Benassayag et al. 2005), suggesting that results found in one system are likely to be generally applicable. To identify genes required for dMyc function in vivo, we carried out a dose-sensitive genetic screen using a rough eye phenotype generated by overexpressing dMyc during eye development. Among the genes identified in the screen is Little imaginal discs (Lid), a Trithorax group (TrxG) protein that we show is a histone demethylase that genetically and physically interacts with Myc to mediate its biological functions.
Results
Overexpression of dMyc during eye development results in a rough eye phenotype
We used the UAS/GAL4 system (Brand and Perrimon 1993) using a glass minimal response element (GMR) Gal4 driver to overexpress dMyc in differentiating neural cells in the eye imaginal disc. This results in adult eyes that are larger than wild type and display a “rough” appearance due to disorganized ommatidia that we have designated as GMM (GMR-Gal4, UAS-dMyc) (Fig. 1). High-magnification views of GMM eyes and computer-assisted measurement of ommatidial areas reveal that their disorganized appearance is due to 33% enlargement of each ommatidium, resulting in disruption of the highly organized ommatidial array (Fig. 1; Supplementary Fig. S1A). This suggests that the GMM phenotype is likely to be due to increased cell growth.
Figure 1.
The GMM phenotype is suppressed by reducing the gene dose of lid. Scanning electron micrographs of adult eyes. (A,D,G) Control GMR-Gal4 flies. (B,E,H) GMM flies. (C,F,I) GMM, lidK6801/+ flies. A–C are 70× magnification, D–F show lateral views (170×), and G–I are 1000× magnification.
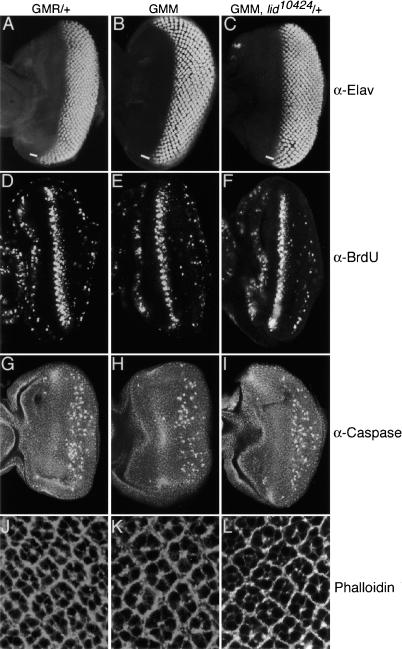
Overexpression of dMyc in wing imaginal disc cells results in accelerated entry of G1 cells into S phase, increased cell mass (larger cells), and an increase in apoptosis (Johnston et al. 1999). In mammalian cells, Myc family genes, in addition to their effects on cell growth, have also been shown to influence cell cycle progression and differentiation (Coppola and Cole 1986; Freytag 1988; Wilson et al. 2004; Okubo et al. 2005). We therefore examined GMM eye discs for effects on differentiation, proliferation, apoptosis, and cell growth. GMM eye discs do not display defects in neuronal differentiation as shown by the expression pattern of Elav, a neural antigen normally expressed in the posterior portion of the eye disc (Fig. 2A,B). Similarly, S phases and apoptosis, detected by BrdU incorporation and expression of cleaved Caspase-3, respectively, are unchanged in GMM eye discs (Fig. 2D,E,G,H). Consistent with the observation that proliferation and apoptosis are unchanged in GMM eye discs, we find no effect on the GMM rough eye phenotype upon reducing the gene dose of cell cycle regulators such as cyclin E, dE2F, or dacapo, or upon deleting the cell death genes hid, grim, and reaper using Df(3L)H99 (White et al. 1996; data not shown). However, monitoring cortical actin using phalloidin staining and analysis of cell size clearly shows that GMM photoreceptor cells are dramatically larger than wild type (Fig. 2J,K; Supplementary Fig. S1B). Thus, dMyc overexpression during eye disc neural differentiation gives rise to an adult rough eye phenotype that is the result of excess cell growth.
Figure 2.
The GMM phenotype is primarily due to increased cell growth. Control GMR-Gal4/+ (A,D,G,J), GMM (B,E,H,K), and GMM, lid10424/+ (C,F,I,L) eye imaginal discs from wandering third instar larvae. (A–C) Anti-Elav to detect differentiating neurons posterior to the morphogenetic furrow (MF; marked with white bar). (D–F) BrdU incorporation to visualize cells in S phase. (G–I) Cell death visualized by expression of cleaved Caspase-3. (J–L) Cortical actin visualized by phalloidin-Rhodamine conjugate. Eye discs are orientated with anterior to the left.
Little imaginal discs (lid) suppresses the GMM rough eye phenotype
Rough eye phenotypes have been used extensively in Drosophila to identify dose-sensitive genetic interactors (Staehling-Hampton et al. 1999; Lane et al. 2000; for review, see St Johnston 2002). To determine whether the GMM phenotype provides a genetically sensitive assay, we tested whether heterozygocity for genes known to positively or negatively regulate dMyc function are capable of suppressing or enhancing the GMM phenotype. Increasing dMyc levels by genetically reducing either Archipelago (Ago), an E3 ubiquitin ligase that mediates dMyc degradation (Moberg et al. 2004) or Hpf, a transcriptional repressor of the dmyc gene, enhances the GMM rough eye phenotype (Quinn et al. 2004; data not shown; Fig. 6D, below). Conversely, the GMM phenotype is suppressed by reducing the levels of the Drosophila homolog of the c-Myc coactivator Pcaf (McMahon et al. 2000; data not shown), confirming the sensitivity of our assay. To identify novel genes that regulate dMyc-regulated cell growth, we crossed the GMM strain to 390 deficiency strains with deletions on the X, 2nd, and 3rd chromosomes that cover >80% of the Drosophila genome. GMM progeny heterozygous for each deficiency were scored for enhancement or suppression of GMM rough eye phenotype.
Figure 6.
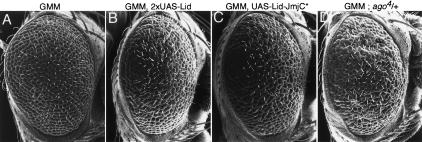
Expression of Lid-JmjC* (full-length Lid protein containing two point mutations in the demethylase domain) enhances the GMM phenotype. (A–E) Scanning electron micrographs of GMM (A), GMM carrying two copies of UAS-Lid (B), GMM, UAS-Lid-JmjC* (C), and GMM; ago4/+ (D). All scanning electron micrographs are 170× magnification.
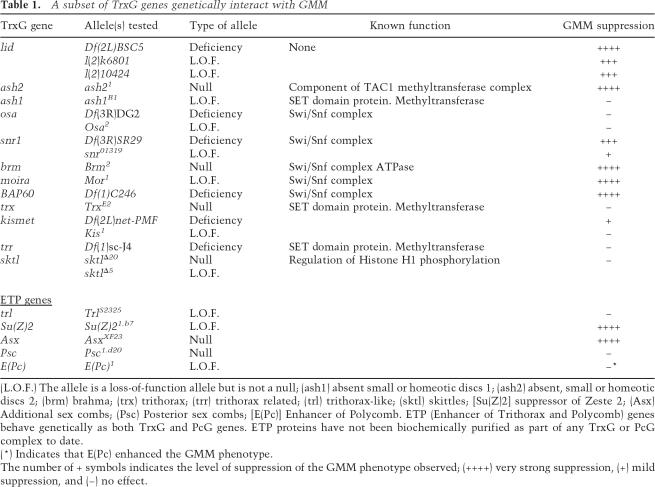
This genetic screen identified 26 suppressor and 15 enhancer regions (to be described elsewhere). One region identified as a strong suppressor of the GMM phenotype was refined to the cytological band 26B1-5 by the region of overlap between the deficiencies Df(2L)E110, Df(2L)BSC5, and Df(2L)cl-h3 (Supplementary Fig. S2A). This region contains ∼14 genes, four of which have characterized mutations. When tested for suppression of GMM, two independently generated P-element insertions, lidk6801 and lid10424, both within the TrxG gene little imaginal discs (lid), were found to suppress the rough eye phenotype by reducing dMyc-induced cell growth (Figs. 1C,F,I, 2; Supplementary Fig. S1A,B; data not shown). The level of suppression observed with these alleles is not as great as with the Df(2L)E110, Df(2L)BSC5, or Df(2L)cl-h3 deletions, consistent with the previously published observation that lidk6801 and lid10424 are hypomorphic alleles (Table 1; data not shown; Gildea et al. 2000). To ensure that the interaction observed between GMM and lid is specific to the growth-inducing function of dMyc and does not reflect a general requirement for Lid in UAS/Gal4-mediated overexpression, both lid alleles were crossed to flies overexpressing Cyclin D/Cdk4 using the GMR-Gal4 driver. Overexpression of Cyclin D/Cdk4 in a wild-type background results in excess cell growth and gives rise to large, bulgy eyes (Datar et al. 2000). Neither lidk6801 nor lid10424 modified this phenotype, demonstrating that Lid is not generally required for either increased cellular growth or for Gal4-mediated transgene expression (data not shown). In addition, the suppression of the GMM phenotype by lidk6801 is specifically due to decreased Lid expression, as reverting this allele to wild type by precisely excising the P-element results in no genetic interaction with GMM (data not shown). Lid is therefore required for dMyc-induced cell growth in the differentiating neurons of the eye imaginal disc.
Table 1.
A subset of TrxG genes genetically interact with GMM
(L.O.F.) The allele is a loss-of-function allele but is not a null; (ash1) absent small or homeotic discs 1; (ash2) absent, small or homeotic discs 2; (brm) brahma; (trx) trithorax; (trr) trithorax related; (trl) trithorax-like; (sktl) skittles; [Su(Z)2] suppressor of Zeste 2; (Asx) Additional sex combs; (Psc) Posterior sex combs; [E(Pc)] Enhancer of Polycomb. ETP (Enhancer of Trithorax and Polycomb) genes behave genetically as both TrxG and PcG genes. ETP proteins have not been biochemically purified as part of any TrxG or PcG complex to date.
(*) Indicates that E(Pc) enhanced the GMM phenotype.
The number of + symbols indicates the level of suppression of the GMM phenotype observed; (++++) very strong suppression, (+) mild suppression, and (−) no effect.
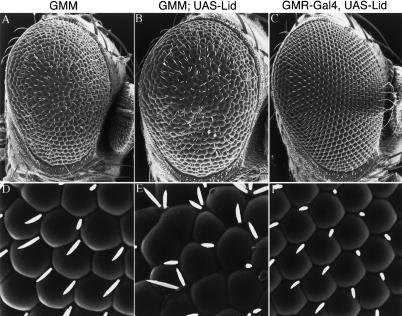
Since reducing Lid levels suppresses the GMM phenotype, we next determined whether overexpression of Lid would enhance GMM. To this end, we generated UAS-Lid transgenic flies to permit Gal4-mediated overexpression of Lid during development. Coexpression of dMyc and Lid using the GMR-Gal4 driver results in a dramatic increase in cell growth and an enhancement of the GMM eye phenotype, suggesting that Lid is limiting for dMyc function (Fig. 3A,B,D,E). Overexpression of Lid alone has no discernable adult eye phenotype, indicating that Lid alone is not sufficient to increase cell growth (Fig. 3C,F). Importantly, Lid does not generally up-regulate Gal4-mediated transcription as UAS-mediated coexpression of Lid, Cyclin D, and Cdk4 showed an eye phenotype identical to that resulting from overexpression of Cyclin D/Cdk4 (GDK) alone (data not shown).
Figure 3.
Increasing Lid levels enhances the GMM phenotype. Scanning electron micrographs of GMM (A,D), GMM, UAS-Lid (B,E), and GMR-Gal4, UAS-Lid (C,F) adult eyes. A–C show lateral views (170× magnification) and D–F show1000× magnification.
We also asked whether suppression of the GMM phenotype, by reducing the gene dose of lid, might be related to an effect of Lid on dmyc mRNA or dMyc protein stability. We therefore examined dMyc levels in immunoblots of wild-type, GMM, and GMM eye discs heterozygous for lid. As shown in Figure 4A, dMyc is significantly overexpressed in GMM eye discs compared with wild type, and this level of expression is unchanged in a lid heterozygous background. Lid is therefore not a regulator of dMyc levels, but is required for dMyc function.
Figure 4.
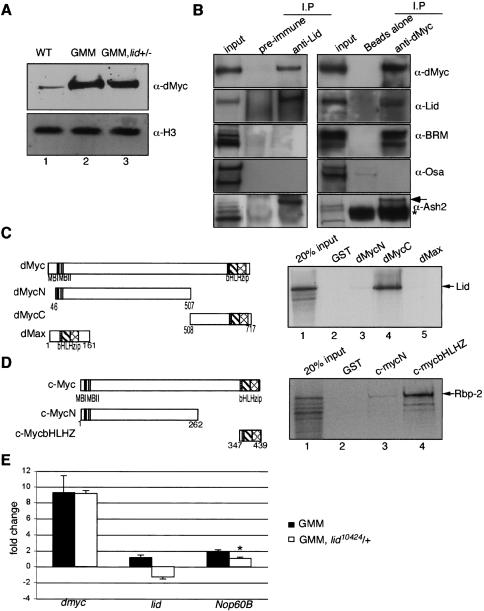
Lid physically interacts with dMyc. (A) Western blot of wild-type (lane 1), GMM (lane 2), and GMM heterozygous for lid10424 (lane 3) using 24 eye discs per lane. Top panel was probed with anti-dMyc; bottom panel shows anti-Histone H3 loading control. (B) Anti-Lid (left panel) and anti-dMyc (right panel) immunoprecipitations and Western blots for dMyc, Lid, Brm, Osa, and Ash2. A high-molecular-weight Ash2 is detected in both anti-Lid and anti-dMyc immunoprecipitates (arrow). (*) A nonspecific band. (C) Lid interacts with the C-terminal region of dMyc in vitro. In vitro assay testing binding of 35S-methionine Lid to GST (lane 2), GST-dMycN (lane 3), GST-dMycC (lane 4), or GST-dMax (lane 5). (D) In vitro binding assay using 35S-methionine-labeled Rbp-2 and GST (lane 2), c-mycN (lane 3), and c-mycbHLHZ (lane 4). (E) Real-time PCR analysis of dmyc, lid, and Nop60B expression levels relative to expression in GMR/+ controls in eye discs from GMM or GMM discs heterozygous for lid10424. Asterisk indicates that Nop60B levels in GMM and GMM heterozygous for lid are significantly different (Student’s t-test; P = 0.003).
lid and dmyc mutants genetically interact
To confirm that Lid is required for endogenous dMyc function, we tested whether reducing the gene dose of lid enhanced the phenotype of a dmyc hypomorphic mutant, dmP0. Male flies hemizygous for dmp0 or other hypomorphic dmyc alleles are viable and fertile but are smaller than wild type and have thin bristles (Gallant et al. 1996; Schreiber-Agus et al. 1997; Johnston et al. 1999). To test for a genetic interaction between lid and dmP0, we used the strong hypomorphic allelic combination of lidk6801 and lid10424 to reduce the gene dose of lid. In control crosses, lidk6801/lid10424 flies eclosed at approximately half the frequency of that expected by Mendelian genetics, and dmP0 hemizygous male flies eclosed at a frequency of 0.8. Of 410 male flies scored, only one dmP0; lidk6801/lid10424 fly was observed, significantly fewer than the 25 expected based on control crosses (P < 0.01) (Table 2). In addition to displaying the small body size and thin bristle phenotypes characteristic of dmP0 males, the dmP0; lidk6801/lid10424 male fly had held-out wings and was sterile. Therefore simultaneously reducing the gene dose of dmyc using dmp0 and lid using lidk6801 and lid10424 reduces fitness. This observation is consistent with Lid being required for endogenous dMyc function in vivo.
Table 2.
lid and dm genetically interact
aNumber of male progeny of the genotypes shown from the cross of w1118, dmP0/w+, dm+; lid10424/CyO females to w67c23; lid6801/CyO males. Animals homozygous for the CyO balancer chromosome do not survive and are not included in the table.
bThe one dmP0; lidk6801/lid10424 male obtained was phenotypically similar to dmP0 males (small, thin bristles), but unlike either dmP0 or lidk6801/lid10424 males, had held-out wings and was sterile.
(*) Indicates that the lid allele can be either lid10424 or lidk6801.
Expected frequencies were generated from two control crosses: (1) w1118, dmP0/w+ females to +/CyO males and (2) lid10424/CyO females to w1118; lid6801/CyO males.
(n.s.) No significant difference between the observed and expected numbers.
Lid and dMyc form a complex in vivo and in vitro
Because TrxG proteins regulate transcription during development (Simon and Tamkun 2002; Brock and Fisher 2005), we considered the possibility that Lid is directly required for dMyc-dependent activation of cell growth genes. If so, then Lid and dMyc would be predicted to form a stable complex in vivo. To test this, we raised rabbit antisera against a bacterially expressed and purified GST-Lid fragment corresponding to amino acids 1427–1665. This anti-Lid antiserum recognizes a single band of ∼200 kDa from Drosophila S2 cell extracts that is consistent with the predicted molecular weight of Lid and is significantly reduced upon treatment with lid-specific RNA interference (RNAi) or in a lid mutant background (Supplementary Fig. S3). Like other Trx-G proteins, Lid is a nuclear protein that is expressed ubiquitously throughout embryonic and larval development (Fig. 5; data not shown). dMyc is detected in anti-Lid immunoprecipitates from third instar larval or S2 tissue culture cell extracts (Fig. 4B; data not shown). Conversely, Lid is also clearly detected in anti-dMyc immunoprecipitates (Fig. 4B). Endogenous Lid and dMyc are therefore stably associated in vivo, a result consistent with our genetic interaction study (see above) implicating Lid in dMyc’s normal physiological function.
Figure 5.
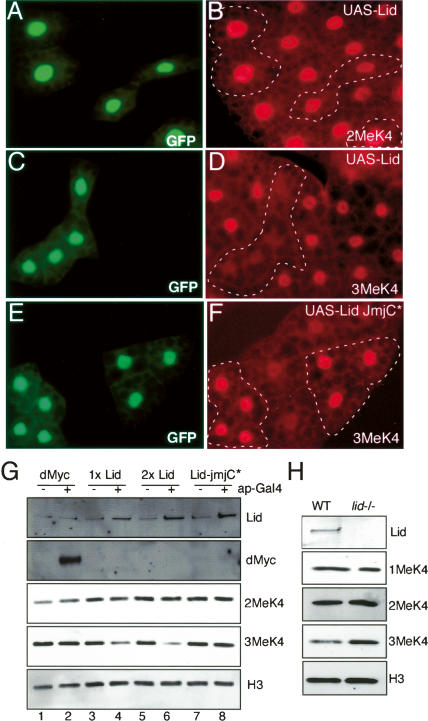
Lid is a trimethyl H3K4 demethylase. (A–F) Flp/Gal4 was used to clonally express Lid (A–D) or Lid-JmjC* (E,F) in fat body cells to examine di- and trimethylated H3K4 levels (red). (A,C,E) Clones expressing Lid or Lid-JmjC* (full-length Lid protein containing two point mutations in the demethylase domain) are marked by coexpression of GFP, and these clones are outlined in the other panels. (G) Western blots from third instar larval wing discs from control (−) or apterous-Gal4 (+) and UAS-dMyc (lanes 1,2), one copy of UAS-Lid (lanes 3,4), two copies of UAS-Lid (lanes 5,6), or UAS-Lid-JmjC* (lanes 7,8). Westerns were probed with anti-Lid, anti-dMyc, anti-dimethyl H3K4, anti-trimethyl H3K4, or a total histone H3 control. Two wing discs were used per lane for all Westerns except 3MeK4, in which four were used. (H) Western analysis of wild-type (left) and lid10424 homozygous mutant (right) wing discs using anti-Lid, anti-monomethyl H3K4, anti-dimethyl H3K4, anti-trimethyl H3K4, or total histone H3.
To map the amino acids required for the dMyc/Lid association, truncated forms of dMyc proteins were tested for their ability to interact with Lid in vitro. GST fusion constructs encoding the N terminus of dMyc (GST-dMycN) containing either the conserved MBI and MBII regions or the C-terminal bHLHZ region required for dimerization and DNA binding (GST-dMycC) were individually tested for their ability to bind in vitro translated, 35S-methionine-labeled, full-length Lid peptide. Lid bound strongly to the C-terminal fragment of dMyc but was not retained by either the GST alone control or GST-dMycN (Fig. 4C). Significantly, the interaction observed between Lid and Myc is evolutionarily conserved, as the human orthologs of Lid, Rbp-2 (JARID1A), and PLU-1 (JARID1B) each bind the bHLHZ region of both c-Myc and N-Myc in parallel GST pull-down assays (Fig. 4D; data not shown). Moreover, we have observed an association between Rbp-2 and c-Myc in mammalian cells (C.-H. Lin, M. Conacci-Sorrell, unpubl.).
A subset of TrxG genes genetically and physically interacts with dMyc
TrxG genes are defined by their ability to genetically enhance other TrxG mutant phenotypes when heterozygous (Simon and Tamkun 2002; Brock and Fisher 2005). In addition to their genetic interactions, TrxG proteins also physically associate to form multiple distinct complexes that alter nucleosome spacing or covalently modify histone tails resulting in context-dependent activation or repression of transcription. To investigate the possibility that other TrxG proteins are similarly involved in dMyc-induced cell growth, TrxG mutants were crossed to GMM flies and the adult eye phenotypes scored (Table 1). GMM flies heterozygous for Su(Z)2, Additional sex combs (Asx), absent small or homeotic discs 2 (ash2), and the snr1, Moira, Brahma, and BAP60 components of the Drosophila Brahma complex (SWI/SNF complex) were dramatically suppressed compared with GMM alone, suggesting that these factors are limiting for dMyc-induced cell growth. Indeed, the level of suppression seen upon reducing the gene dose of Brahma complex components is equivalent to, or better than, that observed when the level of the Drosophila homolog of the established c-Myc coactivator Pcaf is reduced (data not shown).
Lid has not been identified biochemically as a component of any TrxG complex to date. Since subunits of the Brm complex in addition to Lid and Ash2 suppressed the GMM phenotype, these proteins may comprise a novel dMyc transcriptional activation complex. To test this, we immunoprecipitated dMyc or Lid from Drosophila S2 cells and looked for the presence of Brm, Ash2, or Osa. As shown in Figure 4B, Lid, Brm, and Ash2 are coprecipitated with dMyc, while only dMyc and Ash2 are detected in Lid immunoprecipitates. It is interesting to note that a slowly migrating population of Ash2 is the predominant form detected in both anti-dMyc and anti-Lid immunoprecipitates. This high-molecular-weight band is also recognized by an antibody raised to human Ash2 (Ash2L) (data not shown), confirming its identity and suggesting that a covalently modified form of Ash2 is recruited to a dMyc–Lid complex. Consistent with the absence of a genetic interaction between GMM and Osa, we did not detect a physical association of Osa with a dMyc complex. Together, the coimmunoprecipitation results suggest that dMyc forms distinct complexes with TrxG proteins, one with Brm and another that includes Lid and Ash2.
Lid is required for dMyc-dependent activation of Nop60B
To identify genes regulated by the dMyc–Lid complex, we tested the expression of a subset of E-box-containing (Nop5, Nop56, Nop60B, RpI1, RpI135, fibrillarin) and non-E-box-containing (RpIII128 and RpS6) dMyc target genes in GMM eye discs and GMM eye discs heterozygous for lid10424 by real-time PCR. All eight of these genes are up-regulated in response to dMyc overexpression in the wing imaginal disc and whole larvae (Orian et al. 2003; Hulf et al. 2005; data not shown). In contrast, only nucleolar protein at 60B (Nop60B, minifly), an E-box-containing gene essential for ribosomal RNA processing and cell growth (Giordano et al. 1999), is induced in GMM eye discs (Fig. 4E; data not shown), suggesting that not all tissues elicit the same transcriptional program in response to dMyc. Both GMM eye discs and GMM eye discs heterozygous for lid show ninefold higher dmyc mRNA levels than GMR-Gal4 control discs (Fig. 4E). Despite their comparable levels of dmyc expression, Nop60B is up-regulated ∼1.8-fold in GMM eye discs compared with GMM heterozygous for hypomorphic lid. Lid is therefore limiting for dMyc-dependent activation of Nop60B. As expected, GMM eye discs heterozygous for lid10424 have reduced lid transcript levels as a consequence of their reduced lid gene copy number (Fig. 4E). The finding that Lid is required for the expression of a growth regulatory gene is consistent with our initial identification of lid as a suppressor of the GMM phenotype.
Lid encodes a trimethyl histone HK4 demethylase
Lid is a 1838-amino-acid protein possessing numerous conserved motifs including an ARID (A/T-rich interaction domain) (Kortschak et al. 2000), implicated in binding A/T-rich DNA; a single C5HC2 zinc finger; three PHD fingers (plant homeobox domain) (Aasland et al. 1995), implicated in forming protein–protein interactions; and Jumonji N and C (JmjN and JmjC) domains. JmjC-containing proteins have recently been shown to act as histone demethylase enzymes in a Fe2+ and α-ketoglutarate-dependent manner (Klose et al. 2006a). To test whether Lid can demethylate histones in vivo, we overexpressed Lid in fat body and in wing disc cells and examined the levels of mono-, di-, and trimethylated histone H3K4 and H3K27. We also monitored di- and trimethylated histone H4K20 and trimethylated histone H3K9 and H3K36. Overexpression of Lid specifically decreased the levels of the trimethylated form of H3K4 but had no effect on the other methylated histones examined in either GFP-marked fat body or wing disc cells (Fig. 5A–D,G; data not shown). Significantly, expression of Lid in the wing disc reduced trimethyl H3K4 in a dose-dependent manner, with two copies of the UAS-Lid transgene reducing trimethyl H3K4 more efficiently than one copy (Fig. 5G). Moreover, levels of trimethylated H3K4 are increased in wing discs from lid10424 homozygous mutant animals, consistent with the model that Lid regulates the levels of this histone modification during normal development (Fig. 5H). To determine whether the JmjC domain of Lid is required for the observed H3K4 demethylation, we generated transgenic flies carrying a Gal4-inducible form of full-length Lid containing Ala substitutions at His637 and Glu639 (Lid-JmjC*) that abolishes the protein’s ability to bind the Fe2+ cofactor required for demethylase activity. Similar mutations have previously been shown to disrupt the demethylase function of the JmjC domains of JHDM2A, JHDM3A, JHDM1, and JMJD2A (Klose et al. 2006b; Tsukada et al. 2006; Yamane et al. 2006; Whetstine et al. 2006). Unlike wild-type Lid, expression of full-length Lid-JmjC* did not decrease levels of trimethylated H3K4 in fat body or in wing disc cells (Fig. 5E–G), demonstrating that an active JmjC domain is required for Lid-mediated H3K4 demethylation. Interestingly, expression of Lid-JmjC* resulted in increased levels of trimethyl H3K4 in the fat body, perhaps due to a dominant interfering effect on wild-type Lid function in these cells. Taken together, these results demonstrate that Lid is a trimethyl H3K4 demethylase that modifies nucleosomal histone H3 in vivo. The global regulation of H3K4 trimethylation status by Lid is not, however, likely to be mediated by recruitment by dMyc, since we do not observe any effect of reduced or increased dMyc expression on trimethyl H3K4 levels in either fat body or wing disc cells (Fig. 5G; data not shown).
To determine whether Lid’s demethylase activity is required for dMyc function, we crossed 10 independent UAS-Lid-JmjC* lines to GMM and examined their eye phenotypes. To allow a direct comparison between the phenotypes generated by expression of Lid-jmjC* and wild-type Lid, we used a strain carrying two copies of UAS-Lid as this shows similar levels of overexpression to one copy of UAS-Lid-JmjC* (Fig. 5G; data not shown). Expression of two copies of wild-type Lid or one copy of Lid-JmjC* enhanced the GMM phenotype to a similar extent, indicating that the demethylase activity of Lid is not required for dMyc function during eye development (Fig. 6). This enhancement of GMM occurs by increasing cell death rather than by increasing cell size, resulting in a rougher, but not larger, eye phenotype (data not shown). A similar enhancement of the GMM phenotype due to increased apoptosis is observed when dMyc levels are further increased either by adding another UAS-dMyc transgene or by reducing the gene dose of the E3 ligase ago (Fig. 6E; data not shown). This rougher eye phenotype produced by both wild-type Lid and Lid-JmjC* (full-length Lid that lacks demethylase activity) in combination with GMM therefore reflects increased dMyc activity.
dMyc inhibits the demethylase activity of Lid
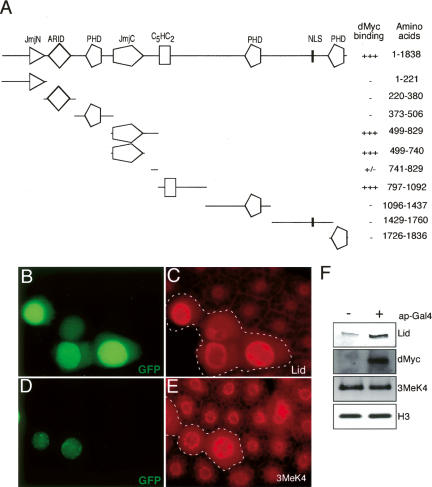
To further investigate the functional interaction between dMyc and Lid, we determined which regions of Lid are required for dMyc binding in vitro. Eight deletion fragments, each containing a conserved domain of Lid, were initially tested for binding to a GST-dMyc fusion protein (summarized in Fig. 7A). Of these eight fragments, two overlapping regions containing the JmjC domain (amino acids 499–829) and the C5HC2 zinc finger (amino acids 797–1092) bound strongly to dMyc. Because these two dMyc-interacting regions overlap by 32 amino acids and could therefore comprise one binding site, we divided the JmjC fragment into JmjC-740 (amino acids 499–740) and a small region that includes the 32-amino-acid overlap (amino acids 741–829) to distinguish their individual binding properties. While the JmjC-740 fragment was sufficient for strong dMyc binding, the region of overlap between the JmjC and C5HC2 zinc finger fragments was not. We therefore conclude that two regions of Lid can bind independently to dMyc—the JmjC demethylase domain (amino acids 499–740) and the JmjC-adjacent fragment that includes the C5HC2 zinc finger (amino acids 797–1092).
Figure 7.
dMyc inhibits the demethylase activity of Lid. (A) Summary of GST fusion protein-mediated in vitro binding assays to map the region of Lid bound by dMyc. (+++) Strong binding (∼20% input); (−) no detectable binding. (B–E) Flp/Gal4-mediated clones coexpressing Lid and dMyc marked by GFP expression (outlined in C,E) and stained for Lid (C) or trimethyl H3K4 (E). (H) Western analysis from UAS-Lid, UAS-dMyc (−), or apterous-Gal4, UAS-Lid, UAS-dMyc (+) wing discs. Westerns were probed with anti-Lid, anti-dMyc, anti-trimethyl H3K4, and total histone H3. Four wing discs were used per lane.
The binding of dMyc to the catalytic JmjC domain of Lid raises the possibility that dMyc may directly inhibit Lid’s demethylase activity or may occlude binding of Lid to its methylated substrate. To test this, we coexpressed dMyc and Lid in vivo and examined trimethylated H3K4 levels. In striking contrast to overexpression of Lid alone, which leads to a dramatic decrease in trimethyl H3K4 levels, no reduction in trimethyl H3K4 is observed when Lid and dMyc are coexpressed in either fat body or wing disc cells (cf. Figs. 7B–F and 5C,D,G). Fat body cells coexpressing dMyc and Lid are, however, significantly larger than their surrounding cells as expected from the induction of a large number of cell growth genes by dMyc (Johnston et al. 1999; Pierce et al. 2004). dMyc therefore functions to inhibit the demethylase activity of Lid.
Discussion
We have identified the TrxG gene lid as a suppressor of the GMM phenotype in a genome-wide genetic screen to identify regulators of dMyc-induced cell growth. In addition to the deletions uncovering lid, we identified 40 other genomic regions that enhanced or suppressed the GMM phenotype when heterozygous. Two of these regions delete genes encoding known regulators of dMyc stability, such as ago, or are involved in Myc transactivation, such as Pcaf. Specific mutations in both of these genes had previously been shown to enhance or suppress the GMM rough eye phenotype, respectively. Interestingly, none of the known direct transcriptional targets of dMyc were identified as genetic modifiers of the GMM phenotype (J. Secombe, unpubl.), suggesting that the GMM phenotype arises from modulation of multiple genes and provides a powerful tool to identify proteins directly required for dMyc function in vivo.
dMyc interacts with Lid and other TrxG proteins
TrxG proteins are renowned for their essential role in maintaining homeotic (hox) gene expression during development, with mutations in many TrxG genes resulting in lethality due to homeotic transformations (Ringrose and Paro 2004). Six TrxG protein complexes have been identified to date. While one function of these complexes is to antagonize Polycomb group (PcG) repression to maintain active hox gene expression, TrxG proteins are also recruited to other developmentally important genes to either activate or repress their transcription in a context-dependent manner (Papoulas et al. 1998; Simon and Tamkun 2002; Angulo et al. 2004; Cheng and Shearn 2004; Mohrmann et al. 2004; Grimaud et al. 2006). Based on the suppression of the GMM phenotype, the physical interaction between Lid and dMyc, and the requirement of Lid for dMyc-dependent activation of Nop60B, we predict that Lid acts as a dMyc coactivator involved in cell growth. The interaction between endogenous Lid and dMyc proteins is also likely to be essential for normal larval development since reducing the gene dose of lid is lethal in combination with the dmyc hypomorphic allele dmP0. In addition, genetically reducing lid enhances a small bristle phenotype induced by expression of a dMyc RNAi transgene (J. Secombe and L. Li, unpubl.). The original small discs phenotype described for lid mutants also suggests a role for Lid in the regulation of cell growth or proliferation during larval development (Gildea et al. 2000). Unfortunately, we find that this phenotype occurs at a frequency far too low (<1% of lid mutant larvae) to allow characterization.
We expect that the function of the Lid–Myc complex is evolutionarily conserved, as the human orthologs of Lid, Rbp-2 (JARID1A) and Plu-1 (JARID1B), bind strongly to c-Myc and dMyc in vitro (Fig. 4D; J. Secombe, unpubl.), and both have been implicated in transcriptional regulation. Originally described as a binding partner for the tumor suppressor protein Retinoblastoma (RB) (Defeo-Jones et al. 1991; Fattaey et al. 1993), Rpb-2 has recently been shown to behave as a coactivator for RB at some promoters while antagonizing RB function at others (Benevolenskaya et al. 2005). Rbp-2 has also been identified as a transcriptional coactivator for nuclear hormone receptors (NRs) (Chan and Hong 2001) and for the LIM domain transcription factor Rhombotin-2 (Mao et al. 1997). In addition, Plu-1 acts as a transcriptional corepressor for BF1 and PAX9 (Lu et al. 1999; Tan et al. 2003). While the transcriptional repression activities of Rbp-2 and Plu-1 are likely to be linked to a conserved trimethyl H3K4 demethylase activity (see below), the molecular mechanism by which they activate transcription remains unclear. We are presently addressing the mechanism by which Lid functions by carrying out genetic screens using phenotypes generated by gain or loss of lid function.
Our coimmunoprecipitation analyses revealed that dMyc is likely to form multiple distinct complexes comprising TrxG proteins: One includes the Brm (SWI/SNF) nucleosome remodeling complex, and another contains Lid and Ash2. Consistent with the physical interaction observed between dMyc and Brm, components of the Brm complex suppress the GMM phenotype when genetically reduced, indicating that they are required for dMyc-induced cell growth. An interaction between Myc and the Brm complex has previously been observed in mammalian cells, where c-Myc interacts with the Brm (Brg1) subunit Ini1, and expression of a dominant-negative Brg1 allele inhibits c-Myc-dependent activation of a synthetic E-box reporter (Cheng et al. 1999). However, the interaction between dMyc and the Brm complex described here using Drosophila provides the first demonstration of a biological significance for this complex.
The second dMyc–TrxG complex we identified includes Lid and Ash2, with Ash2 being immunoprecipitated with both anti-dMyc and anti-Lid antisera. In addition, decreased levels of Ash2 suppress, and increased levels of Ash2 levels enhance, the GMM phenotype (J. Secombe, unpubl.), suggesting that Lid and Ash2 are limiting for dMyc-induced cell growth. In Schizosaccharomyces pombe, the orthologs of Ash2 and Lid (Ash2p and Lid2p) interact in vivo (Roguev et al. 2001, 2003). While Ash2 has no known enzymatic activity, it is an integral component of several conserved complexes, including the SET1 histone methyltransferase complex (TAC1 in Drosophila; MLL in mammals) that is essential for methylation of histone H3K4 (Dou et al. 2006). Biochemical purification of SET1, Lid2p, and Ash2p complexes from S. Pombe has demonstrated that the Lid2p–Ash2p complex is distinct from the SET1–Ash2 complex (Roguev et al. 2003). We found that reducing the gene dose of the SET1 ortholog trx did not affect the GMM phenotype, consistent with the Drosophila Lid–Ash2–dMyc complex also being independent of TAC1 methyltransferase complex (Table 1). The observation that Ash2 is a component of both H3K4 methylating (MLL) and demethylating (Lid) complexes is intriguing and suggests that it may be a crucial modulator of H3K4 methylation status. We are presently testing whether Ash2 is required for Lid-mediated H3K4 demethylation.
Lid is a H3K4 trimethyl-specific demethylase
Lid is the first enzyme characterized that specifically demethylates trimethylated histone H3K4 in vivo. Based on the similarity between Lid and its mammalian orthologs Rbp-2 and Plu-1, we expect this demethylase activity to be conserved. We have demonstrated that the enzymatic activity of Lid requires a functional JmjC domain; however, Lid’s specificity for a trimethylated lysine target is likely to be determined by the presence of a conserved N-terminal JmjN domain. Evidence to date suggests that proteins that possess both a JmjN and a JmjC domain prefer di- or trimethylated lysine substrates, whereas JmjC proteins that lack a JmjN domain demethylate mono- or dimethylated lysines (Cloos et al. 2006; Fodor et al. 2006; Klose et al. 2006b; Whetstine et al. 2006). Indeed, analysis of the crystal structure of JMJD2A, which targets trimethylated H3K9 and K36, has revealed that the JmjN domain makes extensive contacts within the catalytic core of the JmjC domain, presumably accounting for the differences in target specificity between JmjC and JmjN/JmjC proteins (Chen et al. 2006).
Trimethylated H3K4 is often found surrounding the transcriptional start site of active genes (Bernstein et al. 2002, 2005; Santos-Rosa et al. 2002; Schneider et al. 2004) and is strongly correlated with binding by c-Myc (Guccione et al. 2006). The trimethyl H3K4 demethylase activity of Lid would predict that Lid/Rbp-2 proteins may act as transcriptional repressors in a similar manner to LSD1, which demethylates mono- and dimethylated H3K4 (Shi et al. 2004). Consistent with this hypothesis, we observe that a large number of genes are derepressed in microarrays of homozygous lid mutant wing discs (L. Li and J. Secombe, unpubl.). However, we find that expression of dMyc abrogates Lid’s enzymatic activity, indicating that Lid is not acting as a demethylase when bound to dMyc. This is consistent with our observation that expression of the Lid-JmjC* demethylase mutant enhances the GMM eye phenotype. Indeed, Lid behaves as a dMyc coactivator based on the requirement for Lid in dMyc-induced expression of the growth regulator Nop60B (see below). Both activation and repression functions have been previously suggested for Rbp2 (Benevolenskaya et al. 2005). Interestingly, LSD1’s demethylase activity is also negatively regulated by an associated protein, BHC80, in a similar manner to the inhibition of Lid’s enzymatic activity by dMyc (Shi et al. 2004, 2005). Dynamic regulation of histone demethylase activity is therefore likely to be a common feature of regulated gene expression in vivo.
Recently, analysis of c-Myc target gene promoters revealed a strong dependence on trimethylated H3K4 for E-box-dependent c-Myc binding (Guccione et al. 2006). Based on this observation, it is tempting to speculate that although Lid is likely to be enzymatically inactive when complexed with dMyc, Lid may retain its ability to recognize trimethylated H3K4 (perhaps through its JmjN domain) and thereby facilitate appropriate E-box selection. The inhibition of Lid demethylase activity by dMyc may also result in maintenance of local H3K4 trimethylation to permit binding of additional dMyc molecules or other transcription factors. The maintenance of trimethylated H3K4 by dMyc may allow binding of the NURF chromatin remodeling complex that specifically recognizes trimethylated H3K4 (Wysocka et al. 2006). NURF binding, through its large BPTF subunit, has been correlated with spatial control of Hox gene expression and is thought to link H3K4 methylation to ATP-dependent chromatin remodeling (Wysocka et al. 2006). Finally, considering the fact that Lid contains multiple domains potentially involved in DNA binding and protein interaction, it is likely that interaction of Lid/Rbp-2 with Myc in Drosophila and mammalian cells will promote association of other proteins with the Myc–Lid complex, allowing further diversification of Myc function.
Materials and methods
Fly strains
All Drosophila strains used in this study were generated in the Eisenman laboratory or obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu) except the “Drosdel” deficiency collection (B. Edgar, Fred Hutchinson Cancer Research Center, Seattle, WA), the Cyclin E allele dmcycEAR95 (H. Richardson, Peter MacCallum Cancer Centre, Australia), and dap2X10 and ago4 (I. Hariharan, University of California at Berkeley, Berkeley, CA). A precise excision of the lid allele lidk6801 was generated by crossing lidk6801/CyO females to +/CyO; delta2-3, Sb/+ males. Male progeny of the genotype lidk6801/CyO; Δ2-3, Sb/+ were then crossed to Sco/CyO females. From this cross, individual males that no longer had the mini-pw+ eye marker associated with the lidk6801 P element and lacked the Δ2-3, Sb chromosome were selected to generate stocks. Numerous homozygous viable lines were generated in this way, and sequence analysis of the DNA surrounding the P-element insertion site in the homozygous viable line 17.1 verified that this line is derived from a precise excision of lidk6801.
The GMM strain was generated by recombining GMR-Gal4 (2nd chromosome) with UAS-dMyc (2nd chromosome insert) and crossing to a strain with two copies of UAS-dMyc on the 3rd chromosome, resulting in a final genotype of w; GMR-Gal4, UAS-dMyc132/CyO; UAS-dMyc42, UAS-dMyc13 (generated by P. Gallant, University of Zurich, Zurich, Switzerland). The strain overexpressing Cyclin D/Cdk4 under GMR-Gal4 (GDK flies) was obtained from B. Edgar and is described in Datar et al. (2000).
UAS-Lid transgenic flies were generated by cloning the lid ORF into the NotI site of pUASp (Rorth 1998) and injecting it into w1118 embryos by Volodymyr Shcherbatyy (The University of Washington, Seattle, WA). The Lid-jmjC* mutant altering His637 and Glu639 to Ala was generated by site-directed mutagenesis and verified by sequencing. Transgenic flies were generated by BestGene (http://www.thebestgene.com).
DNA clones
A full-length Lid clone in the vector pOT2 (BDGP) was obtained from the Drosophila genome release 2.0 “unigene” set (Rubin et al. 2000). Human JARID1A (RBP-2) was obtained from S.W. Chan (Institute of Molecular and Cell Biology, Singapore) (Chan and Hong 2001). pGEX-dMyc encodes amino acids 46–517 of dMyc and has been described previously (Gallant et al. 1996). pGex-dMycC was generated by subcloning an XhoI–NotI fragment of dMyc encoding amino acids 508–717 into pGEX4T-3. The deletion constructs shown in Figure 7 are all in pCite vector (Invitrogen) and encode the following amino acids: JmjN, 1–221; ARID, 120–380; PHD1, 373–506; JmjC, 499–829; C5HC2, 797–1092; PHD2, 1096–1437; and PHD3, 1429–1838. The JmjC-740 fragment encodes amino acids 499–740 and the fragment between JmjC and C5HC2 encodes amino acids 741–829.
Real-time PCR
Twenty to thirty eye imaginal discs were dissected and placed immediately into Trizol (Invitrogen) to extract RNA followed by RNA clean-up using Qiagen RNeasy. One microgram of total RNA was reverse-transcribed for 1 h at 42°C using SuperScript II Reverse Transcriptase (Invitrogen) and oligo(dT) primer to generate cDNA. Quantitative real-time PCR (qRT-PCR) analysis was performed using SYBR Green I (Molecula Probe). qRT-PCR reactions were performed in duplicate in total volumes of 25 μL containing SYBR Green I PCR Master Mix, 0.5 μg of each gene-specific primer, 2 μL of first-strand cDNA template, and nuclease-free water. All qRT-PCR reactions were performed using the Bio-Rad MyIQ Real-Time PCR system with the following conditions: 5 min at 95°C followed by 40 cycles of denaturation for 30 sec at 95°C, annealing for 30 sec at 62°C, and extension for 1 min at 72°C. Data analysis was performed using MyiQ system software (Bio-Rad). Fold change was calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001). At least three biological replicates were used for each primer set. The primers used were as follows: dmyc, GGCTAGGATAACCC CCAATG and CGCAAGCAAATCGTTAAACA; lid, GCACA TCAATGGAAGGACAA and CTTGAAGCTGGCCACAATCT; Nop60B, GAGTGGCTGACCGGTTATGT and GCTGGAG GTGCTTAACTTGC; Cdc2 (control), GGTGACTCGGAAAT TGACCA and AGCAGGGGAACGTGTTCTTA; and eIF2B (control), GGCCCTGTGGTATCCAACTA and GATCTTCCC CCGCACTAGAT.
Antibodies and immunofluorescence
The anti-Lid rabbit polyclonal antibody 7895J was generated against a GST-Lid fusion protein encoding amino acids 1429–1665. Rabbits were boosted four times before terminal bleed at R and R Rabbitry. α-Lid polyclonal antiserum was used at 1:5000 for Western blots and 1:1000 for tissue staining. Anti-Osa (clone #P3XAg8.653) and rat α-ELAV hybridoma supernatants were obtained from the Developmental Studies Hybridoma Bank (University of Iowa). Anti-cleaved Caspase-3 (Asp175) antibody was obtained from Cell Signaling Technology. Antibodies to mono-, di-, and trimethylated H3K4, H3K27, H4K20, and trimethylated H3K9 were obtained from Upstate Biotechnology and used according to the manufacturer’s specifications. Anti-trimethyl H3K36 was obtained from abcam. α-Ash2 was obtained from A. Shearn (Johns Hopkins University, Baltimore, MD) and the α-Brm antibody was obtained from J. Tamkun (University of California at Santa Cruz, Santa Cruz, CA).
BrdU incorporation and antibody staining of larval discs were carried out according to Secombe et al. (1998) and imaged using a Leica confocal microscope. Propidium iodide staining was carried out as described in Johnston et al. (1999). Alexa Fluor 568-conjugated phalloidin was obtained from Molecular Probes and used at a dilution of 1:50. Somatic clones overexpressing dMyc, Lid, or LidjmjC* marked by the coexpression of GFP were generated as described in Johnston et al. (1999).
Western analysis was carried out using standard protocols, using PVDF membrane (Millipore) and either IgG1-HRP (Zymed) secondary antibody and ECL (Pierce) detection for dMyc or infrared-conjugated secondary antibodies (LiCOR) and Odyssey scanner and software for all other Westerns.
In vitro and in vivo binding assays
In vitro GST fusion protein-binding assays were performed as described in Hurlin et al. (1995). For immunoprecipitations, S2 cells or homogenized third instar larvae were resuspended in 10 mM HEPES (pH 8), 100 mM NaCl, 10% glycerol, 1% NP-40, and complete protease inhibitor cocktail (Roche), incubated on ice for 15 min, then spun to pellet cell debris. Lysate (500 μg) was then used in each immunoprecipitation with anti-Lid antibody or anti-dMyc monoclonal antibody P4C4B10 and protein A or G beads, respectively.
Scanning electron microscopy
Drosophila adult eyes were prepared as described in Datar et al. (2000) and viewed on a JEOL 5800 scanning electron microscope.
Acknowledgments
We thank Bobby Schneider for help with scanning electron microscopy, Aaron Parkhurst for excellent technical help, and Siew Wee Chen, Allen Shearn, and John Tamkun for providing antibodies. We are also grateful to Beth Moorefield, Bruce Edgar, and Steve Henikoff for critical reading of the manuscript, and members of the Edgar and Eisenman laboratories for helpful discussion. This work was supported by grant CA57138 and an American Cancer Society Research Professorship to R.N.E. J.S. was supported by the Lady Tata Memorial Trust (UK).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1523007
References
- Aasland R., Gibson T.J., Stewart A.F., Gibson T.J., Stewart A.F., Stewart A.F. The PHD finger—Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- Adhikary S., Eilers M., Eilers M. Transcriptional regulation and transformation by MYC proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Amati B., Frank S.R., Donjerkovic D., Taubert S., Frank S.R., Donjerkovic D., Taubert S., Donjerkovic D., Taubert S., Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta. 2001;1471:M135–M145. doi: 10.1016/s0304-419x(01)00020-8. [DOI] [PubMed] [Google Scholar]
- Angulo M., Corominas M., Serras F., Corominas M., Serras F., Serras F. Activation and repression activities of ash2 in Drosophila wing imaginal discs. Development. 2004;131:4943–4953. doi: 10.1242/dev.01380. [DOI] [PubMed] [Google Scholar]
- Arabi A., Wu S.Q., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Wu S.Q., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Fahlen S., Hydbring P., Soderberg O., Grummt I., Hydbring P., Soderberg O., Grummt I., Soderberg O., Grummt I., Grummt I., et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Bellosta P., Hulf T., Diop S.B., Usseglio F., Pradel J., Aragnol D., Gallant P., Hulf T., Diop S.B., Usseglio F., Pradel J., Aragnol D., Gallant P., Diop S.B., Usseglio F., Pradel J., Aragnol D., Gallant P., Usseglio F., Pradel J., Aragnol D., Gallant P., Pradel J., Aragnol D., Gallant P., Aragnol D., Gallant P., Gallant P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassayag C., Montero L., Colombie N., Gallant P., Cribbs D., Morello D., Montero L., Colombie N., Gallant P., Cribbs D., Morello D., Colombie N., Gallant P., Cribbs D., Morello D., Gallant P., Cribbs D., Morello D., Cribbs D., Morello D., Morello D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 2005;25:9897–9909. doi: 10.1128/MCB.25.22.9897-9909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya E.V., Murray H.L., Branton P., Young R.A., Kaelin W.G., Murray H.L., Branton P., Young R.A., Kaelin W.G., Branton P., Young R.A., Kaelin W.G., Young R.A., Kaelin W.G., Kaelin W.G. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L., Liu J.S., Kouzarides T., Schreiber S.L., Kouzarides T., Schreiber S.L., Schreiber S.L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Karlsson E.K., Kulbokas E.J., Gingeras T.R., Kulbokas E.J., Gingeras T.R., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Kiermaier A., Dohmann K., Menkel A., Eilers M., Luscher B., Dohmann K., Menkel A., Eilers M., Luscher B., Menkel A., Eilers M., Luscher B., Eilers M., Luscher B., Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes & Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N., Perrimon N. Targeted gene-expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brock H.W., Fisher C.L., Fisher C.L. Maintenance of gene expression patterns. Dev. Dyn. 2005;232:633–655. doi: 10.1002/dvdy.20298. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Hong W.J., Hong W.J. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J. Biol. Chem. 2001;276:28402–28412. doi: 10.1074/jbc.M100313200. [DOI] [PubMed] [Google Scholar]
- Chen Z.Z., Zang J.Y., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Zang J.Y., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Davrazou F., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Kutateladze T.G., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Simpson M., Mao Q.L., Pan C.H., Dai S.D., Mao Q.L., Pan C.H., Dai S.D., Pan C.H., Dai S.D., Dai S.D., et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Cheng M.K., Shearn A., Shearn A. The direct interaction between ASH2, a Drosophila trithorax group protein, and SKTL, a nuclear phosphatidylinositol 4-phosphate 5-kinase, implies a role for phosphatidylinositol 4,5-bisphosphate in maintaining transcriptionally active chromatin. Genetics. 2004;167:1213–1223. doi: 10.1534/genetics.103.018721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.W.G., Davies K.P., Yung E., Beltran R.J., Yu J., Kalpana G.V., Davies K.P., Yung E., Beltran R.J., Yu J., Kalpana G.V., Yung E., Beltran R.J., Yu J., Kalpana G.V., Beltran R.J., Yu J., Kalpana G.V., Yu J., Kalpana G.V., Kalpana G.V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K., Rappsilber J., Antal T., Hansen K.H., Helin K., Antal T., Hansen K.H., Helin K., Hansen K.H., Helin K., Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Cole M.D., Nikiforov M.A., Nikiforov M.A. Transcriptional activation by the Myc oncoprotein. Curr. Top. Microbiol. Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- Coller H.A., Grandori C., Tamayo P., Colbert T., Lander E.S., Eisenman R.N., Golub T.R., Grandori C., Tamayo P., Colbert T., Lander E.S., Eisenman R.N., Golub T.R., Tamayo P., Colbert T., Lander E.S., Eisenman R.N., Golub T.R., Colbert T., Lander E.S., Eisenman R.N., Golub T.R., Lander E.S., Eisenman R.N., Golub T.R., Eisenman R.N., Golub T.R., Golub T.R. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola J.A., Cole M.D., Cole M.D. Constitutive c-myc oncogene expression blocks mouse erythroleukemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Datar S.A., Jacobs H.W., de la Cruz A.F.A., Lehner C.F., Edgar B.A., Jacobs H.W., de la Cruz A.F.A., Lehner C.F., Edgar B.A., de la Cruz A.F.A., Lehner C.F., Edgar B.A., Lehner C.F., Edgar B.A., Edgar B.A. The Drosophila cyclin D-cdk4 complex promotes cellular growth. EMBO J. 2000;19:4543–4554. doi: 10.1093/emboj/19.17.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeo-Jones D., Huang P.S., Jones R.E., Haskell K.M., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A., Huang P.S., Jones R.E., Haskell K.M., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A., Jones R.E., Haskell K.M., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A., Haskell K.M., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A., Hanobik M.G., Huber H.E., Oliff A., Huber H.E., Oliff A., Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- Dou Y.L., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G., Verdine G.L., Allis C.D., Roeder R.G., Allis C.D., Roeder R.G., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R., Farnham P.J., Farnham P.J. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R., Farnham P.J., Farnham P.J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- Fattaey A.R., Helin K., Dembski M.S., Dyson N., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Helin K., Dembski M.S., Dyson N., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Dembski M.S., Dyson N., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Dyson N., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Hanobik M.G., Haskell K.M., Oliff A., Defeojones D., Haskell K.M., Oliff A., Defeojones D., Oliff A., Defeojones D., Defeojones D., et al. Characterization of the retinoblastoma binding-proteins Rbp1 and Rbp2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- Fernandez P.C., Frank S.R., Wang L.Q., Schroeder M., Liu S.X., Greene J., Cocito A., Amati B., Frank S.R., Wang L.Q., Schroeder M., Liu S.X., Greene J., Cocito A., Amati B., Wang L.Q., Schroeder M., Liu S.X., Greene J., Cocito A., Amati B., Schroeder M., Liu S.X., Greene J., Cocito A., Amati B., Liu S.X., Greene J., Cocito A., Amati B., Greene J., Cocito A., Amati B., Cocito A., Amati B., Amati B. Genomic targets of the human c-Myc protein. Genes & Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T., Opravil S., Mechtler K., Schotta G., Jenuwein T., Mechtler K., Schotta G., Jenuwein T., Schotta G., Jenuwein T., Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B., Schroeder M., Fernandez P., Taubert S., Amati B., Fernandez P., Taubert S., Amati B., Taubert S., Amati B., Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H.M., Livingston D.M., Amati B., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H.M., Livingston D.M., Amati B., Taubert S., Fernandez P., Fuchs M., Chan H.M., Livingston D.M., Amati B., Fernandez P., Fuchs M., Chan H.M., Livingston D.M., Amati B., Fuchs M., Chan H.M., Livingston D.M., Amati B., Chan H.M., Livingston D.M., Amati B., Livingston D.M., Amati B., Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag S. Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distict predifferentiation state in G0/G1. Mol. Cell. Biol. 1988;8:1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P. Myc/Max/Mad in invertebrates: The evolution of the Max network. Curr. Top. Microbiol. Immunol. 2006;302:235–253. doi: 10.1007/3-540-32952-8_9. [DOI] [PubMed] [Google Scholar]
- Gallant P., Shiio Y., Cheng P.F., Parkhurst S.M., Eisenman R.N., Shiio Y., Cheng P.F., Parkhurst S.M., Eisenman R.N., Cheng P.F., Parkhurst S.M., Eisenman R.N., Parkhurst S.M., Eisenman R.N., Eisenman R.N. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- Gebhardt A., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M., Benitah S.A., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M., Braun K., Samans B., Watt F.M., Elsasser H.P., Eilers M., Samans B., Watt F.M., Elsasser H.P., Eilers M., Watt F.M., Elsasser H.P., Eilers M., Elsasser H.P., Eilers M., Eilers M. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 2006;172:139–149. doi: 10.1083/jcb.200506057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea J.J., Lopez R., Shearn A., Lopez R., Shearn A., Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano E., Peluso I., Senger S., Furia M., Peluso I., Senger S., Furia M., Senger S., Furia M., Furia M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999;144:1123–1133. doi: 10.1083/jcb.144.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley S.M., James L.P., Eisenman R.N., Cowley S.M., James L.P., Eisenman R.N., James L.P., Eisenman R.N., Eisenman R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J., Ngouenet C., Galloway D.A., Eisenman R.N., White R.J., Galloway D.A., Eisenman R.N., White R.J., Eisenman R.N., White R.J., White R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A., Li L., Orian A., Eisenman R.N., Edgar B.A., Orian A., Eisenman R.N., Edgar B.A., Eisenman R.N., Edgar B.A., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- Grimaud C., Negre N., Cavalli G., Negre N., Cavalli G., Cavalli G. From genetics to epigenetics: The tale of Polycomb group and Trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- Guccione E., Martinato F., Finocchiaro G., Luzi L., Tizzoni L., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Martinato F., Finocchiaro G., Luzi L., Tizzoni L., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Finocchiaro G., Luzi L., Tizzoni L., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Luzi L., Tizzoni L., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Tizzoni L., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Dall’Olio V., Zardo G., Nervi C., Bernard L., Amati B., Zardo G., Nervi C., Bernard L., Amati B., Nervi C., Bernard L., Amati B., Bernard L., Amati B., Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat. Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guo Q.M., Malek R.L., Kim S., Chiao C., He M., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., Malek R.L., Kim S., Chiao C., He M., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., Kim S., Chiao C., He M., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., Chiao C., He M., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., He M., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., Ruffy M., Sanka K., Lee N.H., Dang C.V., Liu E.T., Sanka K., Lee N.H., Dang C.V., Liu E.T., Lee N.H., Dang C.V., Liu E.T., Dang C.V., Liu E.T., Liu E.T. Identification of c-Myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Cordon-Cardo C., Lowe S.W., Hannon G.J., Lowe S.W., Hannon G.J., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Syvaoja J., Saluz H.P., Haenel F., Eilers M., Saluz H.P., Haenel F., Eilers M., Haenel F., Eilers M., Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- Hulf T., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P., Bellosta P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P., Furrer M., Steiger D., Svensson D., Barbour A., Gallant P., Steiger D., Svensson D., Barbour A., Gallant P., Svensson D., Barbour A., Gallant P., Barbour A., Gallant P., Gallant P. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell. Biol. 2005;25:3401–3410. doi: 10.1128/MCB.25.9.3401-3410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin P.J., Foley K.P., Ayer D.E., Eisenman R.N., Hanahan D., Arbeit J.M., Foley K.P., Ayer D.E., Eisenman R.N., Hanahan D., Arbeit J.M., Ayer D.E., Eisenman R.N., Hanahan D., Arbeit J.M., Eisenman R.N., Hanahan D., Arbeit J.M., Hanahan D., Arbeit J.M., Arbeit J.M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P., Edgar B.A., Eisenman R.N., Gallant P., Eisenman R.N., Gallant P., Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A.M., Cole M.D., Rowitch D.H., Cole M.D., Rowitch D.H., Rowitch D.H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D., Adhikary S., Eilers M., Adhikary S., Eilers M., Eilers M. Mechanisms of transcriptional repression by Myc. Curr. Top. Microbiol. Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Kallin E.M., Zhang Y., Kallin E.M., Zhang Y., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006a;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y.J., Zhang D.Z., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Yamane K., Bae Y.J., Zhang D.Z., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Bae Y.J., Zhang D.Z., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Zhang D.Z., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Tempst P., Wong J.M., Zhang Y., Wong J.M., Zhang Y., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006b;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S., Zhang X.Y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Zhang X.Y., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Cheng P.F., Gafken P.R., McMahon S.B., Eisenman R.N., Gafken P.R., McMahon S.B., Eisenman R.N., McMahon S.B., Eisenman R.N., Eisenman R.N. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortschak R.D., Tucker P.W., Saint R., Tucker P.W., Saint R., Saint R. ARID proteins come in from the desert. Trends Biochem. Sci. 2000;25:294–299. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- Lane M.E., Elend M., Heidmann D., Herr A., Marzodko S., Herzig A., Lehner C.F., Elend M., Heidmann D., Herr A., Marzodko S., Herzig A., Lehner C.F., Heidmann D., Herr A., Marzodko S., Herzig A., Lehner C.F., Herr A., Marzodko S., Herzig A., Lehner C.F., Marzodko S., Herzig A., Lehner C.F., Herzig A., Lehner C.F., Lehner C.F. A screen for modifiers of Cyclin E function in Drosophila melanogaster identifies Cdk2 mutations, revealing the insignificance of putative phosphorylation sites in Cdk2. Genetics. 2000;155:233–244. doi: 10.1093/genetics/155.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.R., Van Calcar S., Qu C.X., Cavenee W.K., Zhang M.Q., Ren B., Van Calcar S., Qu C.X., Cavenee W.K., Zhang M.Q., Ren B., Qu C.X., Cavenee W.K., Zhang M.Q., Ren B., Cavenee W.K., Zhang M.Q., Ren B., Zhang M.Q., Ren B., Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Levens D., Levens D. Making Myc. Curr. Top. Microbiol. Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDC method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu P.J., Sundquist K., Baeckstrom D., Poulsom R., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Sundquist K., Baeckstrom D., Poulsom R., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Baeckstrom D., Poulsom R., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Poulsom R., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., Mitchell M., Pitha-Rowe P., Freemont P., Pitha-Rowe P., Freemont P., Freemont P., et al. A novel gene (PLU-1) containing highly conserved putative DNA chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- Luscher B. Function and regulation of the transcription factors of the Mye/Max/Mad network. Gene. 2001;277:1–14. doi: 10.1016/s0378-1119(01)00697-7. [DOI] [PubMed] [Google Scholar]
- Mao S.F., Neale G.A.M., Goorha R.M., Neale G.A.M., Goorha R.M., Goorha R.M. T-cell oncogene rhombotin-2 interacts with retinoblastoma-binding protein 2. Oncogene. 1997;14:1531–1539. doi: 10.1038/sj.onc.1200988. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D., Cole M.D., Van Buskirk H.A., Dugan K.A., Copeland T.D., Cole M.D., Dugan K.A., Copeland T.D., Cole M.D., Copeland T.D., Cole M.D., Cole M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Wood M.A., Cole M.D., Wood M.A., Cole M.D., Cole M.D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg K.H., Mukherjee A., Veraksa A., Artavanis-Tsakonas S., Hariharan I.K., Mukherjee A., Veraksa A., Artavanis-Tsakonas S., Hariharan I.K., Veraksa A., Artavanis-Tsakonas S., Hariharan I.K., Artavanis-Tsakonas S., Hariharan I.K., Hariharan I.K. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 2004;14:965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Mohrmann L., Langenberg K., Krijgsveld J., Kal A.J., Heck A.J.R., Verrijzer C.P., Langenberg K., Krijgsveld J., Kal A.J., Heck A.J.R., Verrijzer C.P., Krijgsveld J., Kal A.J., Heck A.J.R., Verrijzer C.P., Kal A.J., Heck A.J.R., Verrijzer C.P., Heck A.J.R., Verrijzer C.P., Verrijzer C.P. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol. Cell. Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov M.A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B., Cole M.D., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B., Cole M.D., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B., Cole M.D., Kotenko I., Matheos D., Johnsson A., McMahon S.B., Cole M.D., Matheos D., Johnsson A., McMahon S.B., Cole M.D., Johnsson A., McMahon S.B., Cole M.D., McMahon S.B., Cole M.D., Cole M.D. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell B.C., Cheung A.F., Simkevich C.P., Tam W., Ren X.J., Mateyak M.K., Sedivy J.M., Cheung A.F., Simkevich C.P., Tam W., Ren X.J., Mateyak M.K., Sedivy J.M., Simkevich C.P., Tam W., Ren X.J., Mateyak M.K., Sedivy J.M., Tam W., Ren X.J., Mateyak M.K., Sedivy J.M., Ren X.J., Mateyak M.K., Sedivy J.M., Mateyak M.K., Sedivy J.M., Sedivy J.M. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 2003;278:12563–12573. doi: 10.1074/jbc.M210462200. [DOI] [PubMed] [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Zeller K.I., Dang C.V., Mendell J.T., Dang C.V., Mendell J.T., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okubo T., Knoepfler P.S., Eisenman R.N., Hogan B.L., Knoepfler P.S., Eisenman R.N., Hogan B.L., Eisenman R.N., Hogan B.L., Hogan B.L. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132:1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., van Steensel B., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., Delrow J., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., Bussemaker H.J., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., Li L., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., Sawado T., Williams E., Loo L.W.M., Cowley S.M., Yost C., Williams E., Loo L.W.M., Cowley S.M., Yost C., Loo L.W.M., Cowley S.M., Yost C., Cowley S.M., Yost C., Yost C., et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A., Grewal S.S., Knoepfler P.S., Edgar B.A., Parkhurst S.M., Eisenman R.N., Grewal S.S., Knoepfler P.S., Edgar B.A., Parkhurst S.M., Eisenman R.N., Knoepfler P.S., Edgar B.A., Parkhurst S.M., Eisenman R.N., Edgar B.A., Parkhurst S.M., Eisenman R.N., Parkhurst S.M., Eisenman R.N., Eisenman R.N. Genomic binding and transcriptional regulation by the Drosophila myc and mnt transcription factors. Cold Spring Harb. Symp. Quant. Biol. 2005;70:299–307. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- Oskarsson T., Trumpp A., Trumpp A. The Myc trilogy: Lord of RNA polymerases. Nat. Cell Biol. 2005;7:215–217. doi: 10.1038/ncb0305-215. [DOI] [PubMed] [Google Scholar]
- Oster S.K., Ho C.S.W., Soucie E.L., Penn L.Z., Ho C.S.W., Soucie E.L., Penn L.Z., Soucie E.L., Penn L.Z., Penn L.Z. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- Papoulas O., Beek S.J., Moseley S.L., McCallum C.M., Sarte M., Shearn A., Tamkun J.W., Beek S.J., Moseley S.L., McCallum C.M., Sarte M., Shearn A., Tamkun J.W., Moseley S.L., McCallum C.M., Sarte M., Shearn A., Tamkun J.W., McCallum C.M., Sarte M., Shearn A., Tamkun J.W., Sarte M., Shearn A., Tamkun J.W., Shearn A., Tamkun J.W., Tamkun J.W. The Drosophila Trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- Patel J.H., Loboda A.P., Showe M.K., Showe L.C., McMahon S.B., Loboda A.P., Showe M.K., Showe L.C., McMahon S.B., Showe M.K., Showe L.C., McMahon S.B., Showe L.C., McMahon S.B., McMahon S.B. Opinion—Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Pierce S.B., Yost C., Britton J.S., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N., Yost C., Britton J.S., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N., Britton J.S., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N., Flynn E.M., Edgar B.A., Eisenman R.N., Edgar B.A., Eisenman R.N., Eisenman R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131:2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- Quinn L.M., Dickins R.A., Coombe M., Hime G.R., Bowtell D.D.L., Richardson H., Dickins R.A., Coombe M., Hime G.R., Bowtell D.D.L., Richardson H., Coombe M., Hime G.R., Bowtell D.D.L., Richardson H., Hime G.R., Bowtell D.D.L., Richardson H., Bowtell D.D.L., Richardson H., Richardson H. Drosophila Hfp negatively regulates dmyc and stg to inhibit cell proliferation. Development. 2004;131:1411–1423. doi: 10.1242/dev.01019. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Pijnappel W.W.M.P., Wilm M., Aasland R., Stewart A.F., Schaft D., Shevchenko A., Pijnappel W.W.M.P., Wilm M., Aasland R., Stewart A.F., Shevchenko A., Pijnappel W.W.M.P., Wilm M., Aasland R., Stewart A.F., Pijnappel W.W.M.P., Wilm M., Aasland R., Stewart A.F., Wilm M., Aasland R., Stewart A.F., Aasland R., Stewart A.F., Stewart A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Aasland R., Shevehenko A., Stewart A.F., Schaft D., Shevchenko A., Aasland R., Shevehenko A., Stewart A.F., Shevchenko A., Aasland R., Shevehenko A., Stewart A.F., Aasland R., Shevehenko A., Stewart A.F., Shevehenko A., Stewart A.F., Stewart A.F. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 2003;278:8487–8493. doi: 10.1074/jbc.M209562200. [DOI] [PubMed] [Google Scholar]
- Rorth P. Gal-4 in the Drosophila germline. Mech. Dev. 1998;78:113–118. doi: 10.1016/s0925-4773(98)00157-9. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Hong L., Brokstein P., Evans-Holm M., Frise E., Stapleton M., Harvey D.A., Hong L., Brokstein P., Evans-Holm M., Frise E., Stapleton M., Harvey D.A., Brokstein P., Evans-Holm M., Frise E., Stapleton M., Harvey D.A., Evans-Holm M., Frise E., Stapleton M., Harvey D.A., Frise E., Stapleton M., Harvey D.A., Stapleton M., Harvey D.A., Harvey D.A. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T., Schreiber S.L., Mellor J., Kouzarides T., Mellor J., Kouzarides T., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T., Thorne A.W., Crane-Robinson C., Kouzarides T., Crane-Robinson C., Kouzarides T., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N., Stein D., Chen K., Goltz J.S., Stevens L., DePinho R.A., Stein D., Chen K., Goltz J.S., Stevens L., DePinho R.A., Chen K., Goltz J.S., Stevens L., DePinho R.A., Goltz J.S., Stevens L., DePinho R.A., Stevens L., DePinho R.A., DePinho R.A. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc. Natl. Acad. Sci. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M., Kohlhuber F., Holzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Kohlhuber F., Holzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Holzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Laux G., Polack A., Weidle U.H., Polack A., Weidle U.H., Weidle U.H., et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J., Pispa J., Saint R., Richardson H., Pispa J., Saint R., Richardson H., Saint R., Richardson H., Richardson H. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for Cyclin E in cell proliferation control during eye imaginal disc development. Genetics. 1998;149:1867–1882. doi: 10.1093/genetics/149.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massague J., Pouponnot C., Staller P., Schader M., Eilers M., Massague J., Staller P., Schader M., Eilers M., Massague J., Schader M., Eilers M., Massague J., Eilers M., Massague J., Massague J. TGF β influences Myc, Miz-1 and Smad to control the CDK inhibitor p15(INK4b) Nat. Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- Shen X.T., Mizuguchi G., Hamiche A., Wu C., Mizuguchi G., Hamiche A., Wu C., Hamiche A., Wu C., Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shi Y.J., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Whetstine J.R., Cole P.A., Casero R.A., Shi Y., Cole P.A., Casero R.A., Shi Y., Casero R.A., Shi Y., Shi Y. Histone demethylation mediated by the nuclear arnine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y.J., Matson C., Lan F., Iwase S., Baba T., Shi Y., Matson C., Lan F., Iwase S., Baba T., Shi Y., Lan F., Iwase S., Baba T., Shi Y., Iwase S., Baba T., Shi Y., Baba T., Shi Y., Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Simon J.A., Tamkun J.W., Tamkun J.W. Programming off and on states in chromatin: Mechanisms of Polycomb and Trithorax group complexes. Curr. Opin. Genet. Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Ciampa P.J., Brook A., Dyson N., Ciampa P.J., Brook A., Dyson N., Brook A., Dyson N., Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Kiermaier A., Seoane J., Lukas J., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Seoane J., Lukas J., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Lukas J., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Karsunky H., Moroy T., Bartek J., Massague J., Hanel F., Moroy T., Bartek J., Massague J., Hanel F., Bartek J., Massague J., Hanel F., Massague J., Hanel F., Hanel F., et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 2002;3:176–178. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- Tan K., Shaw A.L., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P.S., Shaw A.L., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P.S., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P.S., Jensen K., Taylor-Papadimitriou J., Freemont P.S., Taylor-Papadimitriou J., Freemont P.S., Freemont P.S. Human PLU-1 has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 2003;278:20507–20513. doi: 10.1074/jbc.M301994200. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M., Gasser S., Murphy M., Martin G.R., Bishop J.M., Murphy M., Martin G.R., Bishop J.M., Martin G.R., Bishop J.M., Bishop J.M. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Warren M.E., Borchers C.H., Tempst P., Zhang Y., Borchers C.H., Tempst P., Zhang Y., Tempst P., Zhang Y., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Lan F., Huarte M., Smolikov S., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Huarte M., Smolikov S., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Smolikov S., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Chen Z.Z., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Spooner E., Li E., Zhang G.Y., Colaiacovo M., Li E., Zhang G.Y., Colaiacovo M., Zhang G.Y., Colaiacovo M., Colaiacovo M., et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- White K., Tahaoglu E., Steller H., Tahaoglu E., Steller H., Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A., Knabenhans C., Macdonald H.R., Trumpp A., Macdonald H.R., Trumpp A., Trumpp A. c-Myc controls the balance between hematopoietic stem cell self renewal and differentiation. Genes & Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.A., McMahon S.B., Cole M.D., McMahon S.B., Cole M.D., Cole M.D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Tackett A.J., Chait B.T., Badenhorst P., Chait B.T., Badenhorst P., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Erdjument-Bromage H., Tempst P., Wong J.M., Zhang Y., Tempst P., Wong J.M., Zhang Y., Wong J.M., Zhang Y., Zhang Y. JHDM2A, a JmjC-containing H3K9 dernethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Chasse D., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Monteith T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Ivaldi G., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Hahn W.C., Stukenberg P.T., Shenolikar S., Uchida T., Stukenberg P.T., Shenolikar S., Uchida T., Shenolikar S., Uchida T., Uchida T., et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]