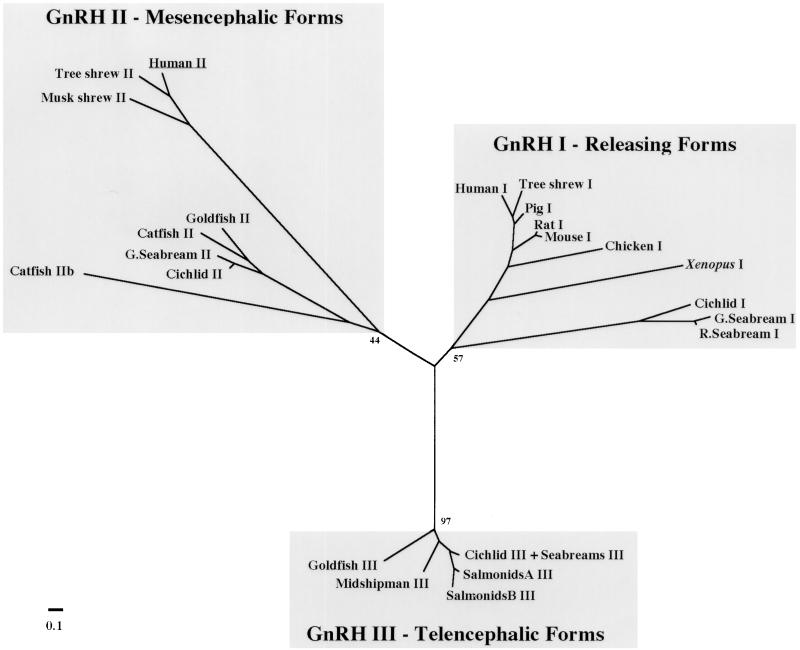
Figure 4.
Unrooted neighbor-joining phylogenetic tree of GnRH peptides. The tree was generated from an alignment of GnRH preprohormones by using the neighbor-joining algorithm (11) and the Hall distance calculation correction procedure in phylip (10). Evolutionary distances are represented only by the length of the branches and not by branch angles. Bootstrap values, indicating the number of times a particular set of sequences group together when trees are generated from resampled alignments, are indicated for some important nodes on the tree. An unrooted tree is shown because of the lack of an obvious outgroup. Some GnRH-III peptide sequences from different species were identical and are grouped together. The scale bar corresponds to estimated evolutionary distance units as calculated by protdist in phylip. For accession numbers and species names, see Materials and Methods.