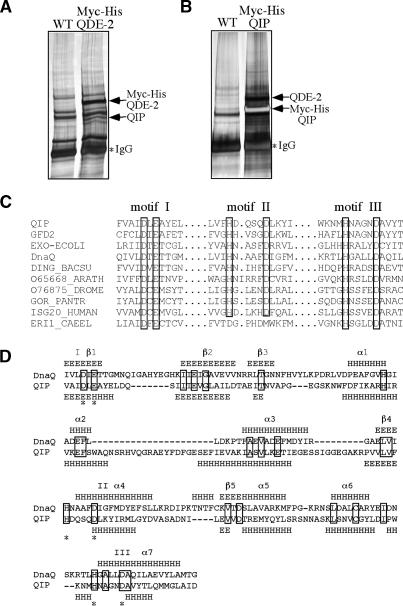
Figure 2.
Identification of QIP as a QDE-2-interacting protein with an exonuclease domain. (A,B) Silver-stained SDS-PAGE gels showing the final immunoprecipitation products of the Myc-His-QDE-2 strain (A) and the Myc-His-QIP strain (B) by c-Myc monoclonal antibody. The identity of the indicated proteins was confirmed by mass spectrometry analyses. (C) Amino acid alignment of the three critical motifs in the exonuclease domains in QIP, ERI-1, and other proteins, including GFD2, Saccharomyces cerevisiae, NP_009894; EXO-ECOLI, E. coli, P0AEK0; DnaQ, E. coli, P03007; DING_BACSU, Bacillus subtillis, P54394; O65668_ARATH, Arabidopsis thaliana, O65668; O76875_DROME, Drosophila melanogaster, O76875; GOR_PANTER, Pan troglodytes, P48778; ISG20_HUMAN, Homo sapiens, Q96AZ6; ERI1_CAEEL, C. elegans, O44406. (D) Secondary structure comparison between the exonuclease domains in QIP and DnaQ (E. coli DNA polymerase III ε subunit). The predicted secondary structure of QIP was obtained by JPred (http://www.compbio.dundee.ac.uk/~www-jpred), whereas the secondary structure of DnaQ was based on its crystal structure (Hamdan et al. 2002).