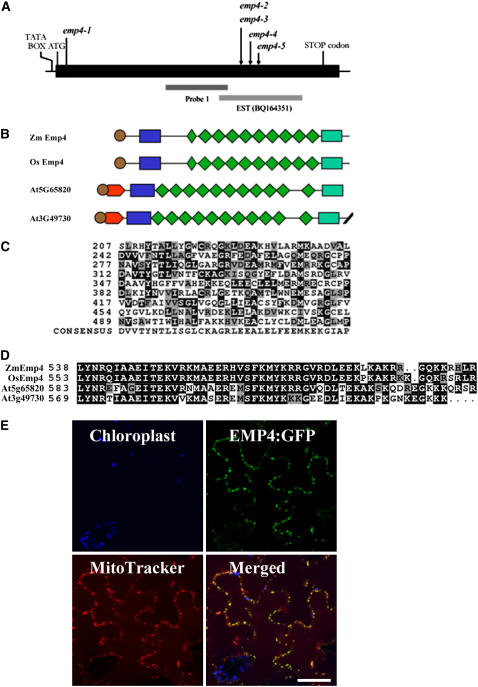
Figure 7.
Organization of emp4 Gene and Protein Structure.
(A) Scheme of the emp4 gene. The location of the Mu3 transposon insertion of the emp4-1 allele in the coding sequence and the positions of Mu insertions of known emp4 alleles are indicated with arrows. The putative TATA box, ATG transcriptional start site, and stop codon are shown. Genomic regions comprising probe 1 and EST clone BQ164351 are indicated.
(B) Schematic alignments of maize emp4 (Zm EMP4), rice emp4-like (Os EMP4; AC135956), and Arabidopsis At3g49730 and At5g65820 predicted amino acid products. The PPR motifs and the PPR-like short motif are represented by green rhomboids. The putative signal peptide is shown as brown circles. The novel N-terminal and C-terminal domains are indicated as blue and green boxes, respectively.
(C) Comparison of the nine PPR motifs found in EMP4 with the PPR consensus sequence. Residues identical to the consensus are shaded in black, and similar residues are shaded in gray.
(D) Alignment of the novel EMP4 C-terminal domain present in Zm Emp4, rice Os Emp4 (AC135956), and Arabidopsis At3g49730 and At5g65820.
(E) EMP4 localization in tobacco leaf epidermal cells. Panels show chloroplast autofluorescence (blue), transient expression of EMP4-GFP (green), Mito Tracker Orange localization (red), and merged images. Fluorescent signals were visualized using confocal laser scanning microscopy.