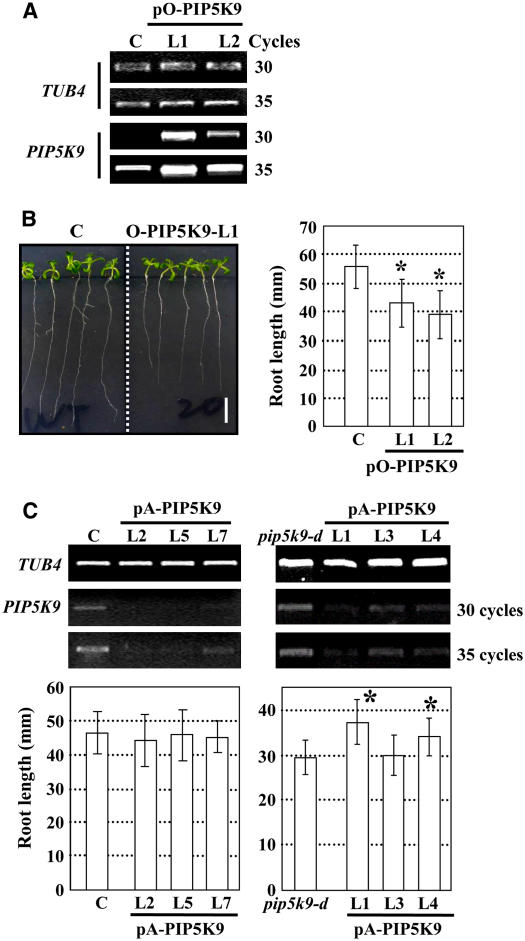
Figure 3.
Analysis of Transgenic Plants Overexpressing or Deficient in PIP5K9.
(A) RT-PCR analysis showed increased transcript levels of PIP5K9 in transgenic lines 5K9-O-L1 and -L2, transformed with pO-PIP5K9. Arabidopsis TUB4 was used as a positive internal control.
(B) Transgenic plants overexpressing PIP5K9 showed shorter roots compared with control plants (C) (left panel; bar = 10 mm). Calculation and statistical heteroscedastic t test analysis showed a significant decrease of root lengths of the PIP5K9-overexpressing plants (*P < 0.01). Twelve-day-old seedlings were used for root length measurement. Error bars represent se (n > 30).
(C) RT-PCR analysis of PIP5K9 transcript levels in control (C) or pip5k9-d mutant plants transformed with pA-PIP5K9 (top panel). Expression of PIP5K9 was repressed in both control and pip5k9-d plants. The At TUB4 gene was used as a positive internal control. Further analysis showed that repression of PIP5K9 in control plants exhibited no obvious difference in root growth, whereas pip5k9-d plants recovered the root length to some extent. Twelve-day-old plants were measured and analyzed. Error bars represent se (n > 30). Heteroscedastic t test analysis showed a significant difference (*P < 0.01).