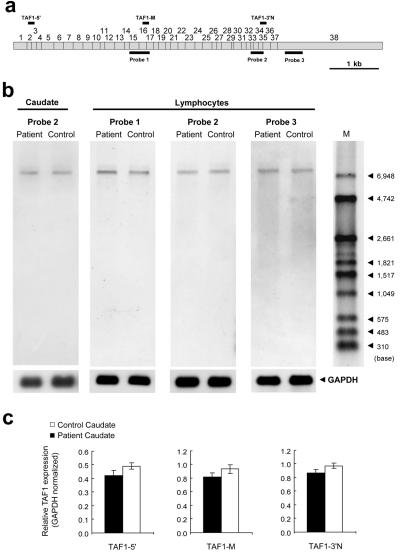
Figure 2. .
Northern analysis. a, Three probes for northern hybridization to TAF1 (detailed information on these probes is given in table 3). b, Total RNA samples from the caudate and lymphoblastoid tissues. The hybridization signal seen at ∼7 kb, which represents TAF1, was observed in every lane, but a signal was never seen at the smaller sizes corresponding to MTS transcripts reported elsewhere. This result suggests that the sequences of TAF1 isoforms have such small differences in size, probably less than a few hundred base pairs, that standard northern analysis cannot discriminate between them in 1% agarose gels denatured by 2% formaldehyde. A probe for GAPDH was also hybridized to each membrane as a loading control. M = DIG-labeled molecular-weight marker. c, Relative TAF1 expression. The hybridization signal seen at ∼7 kb, representing TAF1, had a tendency toward slight reduction in patient caudate, so, to confirm this, TaqMan assays were performed.