Figure 1.
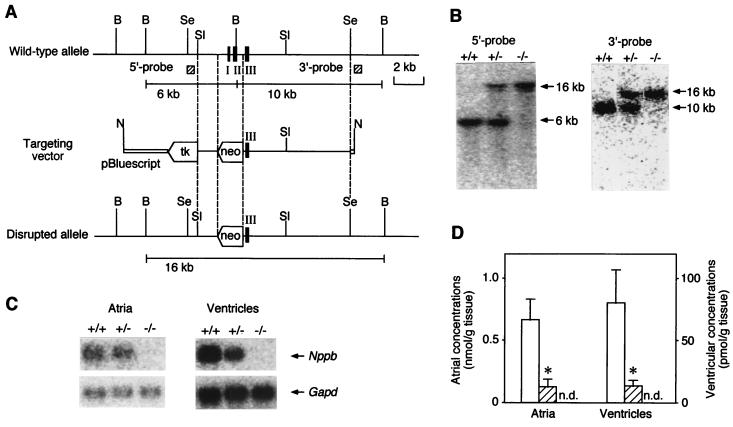
Targeted disruption of Nppb. (A) Restriction maps of the wild-type 129/Sv mouse Nppb allele, targeting vector, and predicted disrupted allele. Filled bars indicate exons (I–III). The genomic fragments used as 5′ and 3′ external probes are shown as hatched boxes. B, BamHI; Se, SpeI; Sl, SalI; N, NotI; tk, herpes simplex virus thymidine kinase gene; neo, neomycin-resistance gene. (B) Southern blot analysis of genomic DNAs from F2 offspring generated by heterozygous intercrosses of F1 mice upon digestion with BamHI. (C) Northern blot analysis of Nppb mRNA in atria and ventricles from Nppb+/+, Nppb+/−, and Nppb−/− mice. Total RNAs (1 and 10 μg) from atria and ventricles, respectively, were analyzed. (D) Cardiac BNP concentrations in Nppb+/+, Nppb+/−, and Nppb−/− mice (n = 3). Open and hatched bars represent Nppb+/+ and Nppb+/− mice, respectively. n.d., not detectable in Nppb−/− mice. ∗, P < 0.05 vs. Nppb+/+ mice.