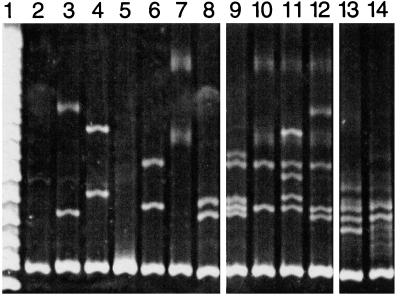
Figure 2.
Heteroduplex mobility assay (HMA) of dinoflagellate SSU rDNA sequences assayed against G. sanguineum “driver” DNA. Assayed “target” and driver DNAs were amplified with the primer pair (dino/4618); all lanes contain the same driver DNA, with added target sequences as indicated. Lanes: 1, DNA ladder; 2, G. sanguineum driver DNA alone; 3, Gymnodinium sp.; 4, P. minimum; 5, King's Creek, MD uncharacterized dinoflagellate DNA clone; 6, Pocomoke River, MD, uncharacterized dinoflagellate DNA clone; 7, uncharacterized dinoflagellate DNA clone derived from a lesioned menhaden (Brevoortia tyrannus); 8, P. piscicida; 9–12, intentionally mixed DNA(s) from lanes 3–8, demonstrating the ability of the assay to simultaneously detect multiple templates; 9, lanes 6 and 8; 10, lanes 6 and 7; 11, lanes 4, 6, and 7; 12, lanes 3, 4, 6, and 7 (the electrophoretic mobility of individual heteroduplex strands is not altered in the presence of mixed amplicons, but the overall HMA pattern become increasingly complex as potential strand annealing combinations multiply with increasing template DNA heterogeneity. With increasing complexity, some heteroduplex bands are lost); 13 and 14, dinoflagellate DNA derived from estuarine water samples (13, King's Creek, MD; 14, Pocomoke River, MD) demonstrating dinoflagellate diversity present in those samples at that time.