Figure 5.
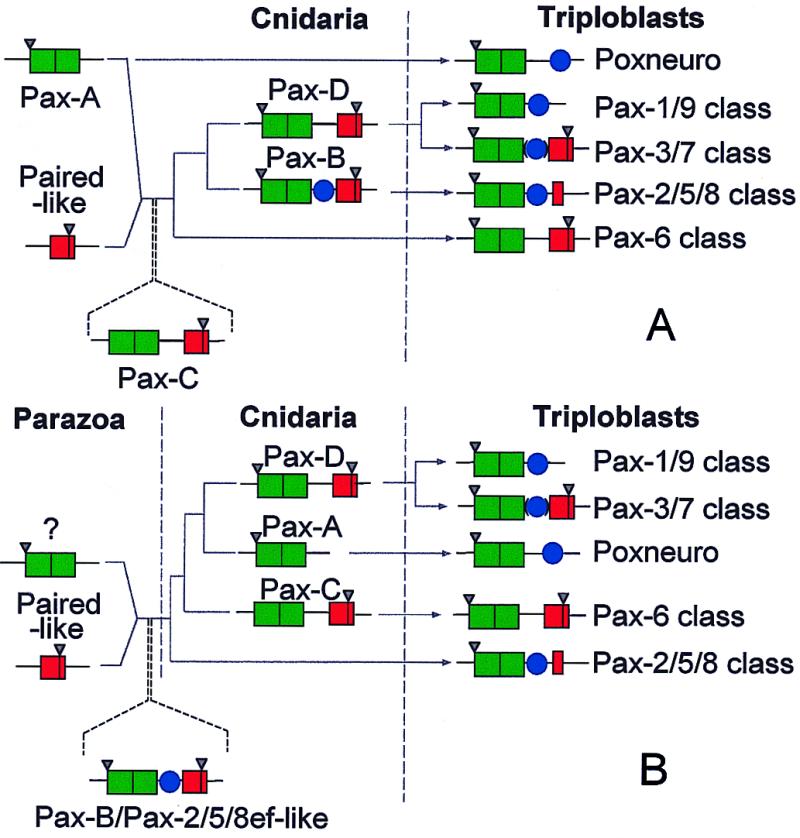
Alternative models for evolution of the Pax classes. (A) This model is based on our previous scheme (18) but accommodates the data reported in this study. Herein, we view Pax-Aam and Pax-Cam as representing the before and after states of a fusion event between a Pax-A/Pox-n-like gene and a paired-like homeobox gene. (B) An alternative model, in which Pax-B/sPax-2/5/8 (rather than Pax-Cam) is seen as reflecting the ancestor of most of the metazoan Pax genes. The models also differ in interpreting the Pax-A/Pox-n class as ancestral (A) or derived (B). In both schemes, Pax-Cam corresponds to the ancestor of the Pax-6 class, and Pax-Dam the precursor of the Pax-3/7 class; the Pax-1/9 class is related to the Pax-3/7 class via loss of the homeobox. Common ancestry of Pax genes is implied by the fact that all of the cnidarian Pax HDs are intermediate between those of the Pax-6 and Pax-3/7 classes (although that in Pax-Dam is very close to the latter class) and share splice sites. Pax-Aam, Pax-Bam, and Pax-Dam have an intron at positions corresponding to the first codon of the PD that is also present in a number of other Pax genes, and each of the Acropora Pax homeoboxes contains an intron at a position corresponding to amino acids 46/47 of the HD (these are indicated by inverted triangles in the scheme). Green box, PD; red box, HD; blue circle, octapeptide.
