Abstract
Genomic technologies have the potential to greatly increase the efficiency of the drug development process. As part of our tuberculosis drug discovery program, we used DNA microarray technology to profile drug-induced effects in Mycobacterium tuberculosis. Expression profiles of M. tuberculosis treated with compounds that inhibit key metabolic pathways are required as references for the assessment of novel antimycobacterial agents. We have studied the response of M. tuberculosis to treatment with the mycolic acid biosynthesis inhibitors isoniazid, thiolactomycin, and triclosan. Thiolactomycin targets the β-ketoacyl-acyl carrier protein (ACP) synthases KasA and KasB, while triclosan inhibits the enoyl-ACP reductase InhA. However, controversy surrounds the precise mode of action of isoniazid, with both InhA and KasA having been proposed as the primary target. We have shown that although the global response profiles of isoniazid and thiolactomycin are more closely related to each other than to that of triclosan, there are differences that distinguish the mode of action of these two drugs. In addition, we have identified two groups of genes, possibly forming efflux and detoxification systems, through which M. tuberculosis may limit the effects of triclosan. We have developed a mathematical model, based on the expression of 21 genes, which is able to perfectly discriminate between isoniazid-, thiolactomycin-, or triclosan-treated M. tuberculosis. This model is likely to prove invaluable as a tool to improve the efficiency of our drug development programs by providing a means to rapidly confirm the mode of action of thiolactomycin analogues or novel InhA inhibitors as well as helping to translate enzyme activity into whole-cell activity.
There is an urgent need for novel drugs that are active against Mycobacterium tuberculosis in order to shorten the duration of tuberculosis therapy and help prevent relapse (24). The sequencing of the complete genome of M. tuberculosis (11) has provided an invaluable resource in the search for such novel antimycobacterial therapies and has facilitated functional genomic studies such as transcriptomics and proteomics that are capable of investigating the biology of M. tuberculosis at the whole-genome level (5). The use of DNA microarrays is one such approach and has many applications within the drug discovery process, from target identification and validation through hit generation and lead optimization (4).
An important factor in developing a new drug is the criterion that the compound must provide selective inhibition of the intended target. Microarray profiling may be useful in delivering this confirmation, through comparison of gene expression profiles obtained in response to novel inhibitors with signature gene expression profiles of drugs of known modes of action. This approach has been demonstrated in Saccharomyces cerevisiae, for which the mode of action of a novel inhibitor was predicted through the concordance of responsive genes with drugs known to inhibit ergosterol biosynthesis (2). Transcript profiling of the response of Haemophilus influenzae to the DNA gyrase inhibitors novobiocin and ciprofloxacin demonstrated that the two different modes of action were clearly reflected in the cellular response (14). In a more recent study, signature profiles generated in response to treatment of Streptococcus pneumoniae with translation inhibitors enabled distinction among antibiotics inhibiting different steps in the translation cycle (23). The response of M. tuberculosis to treatment with the antimycobacterial drugs isoniazid and ethionamide has also been studied (33). Induced genes could be predicted to either compensate for inhibition of the target pathway or respond to the toxic effect of the drug and led to the proposal that RNA response profiles could serve as a fingerprint of a given drug's mode of action (33).
Isoniazid, thiolactomycin, and triclosan all inhibit the biosynthesis of mycolic acids, one of the major components of the mycobacterial cell wall (9), but have different mechanisms of action (17). Isoniazid is a front-line antimycobacterial drug that targets the type II fatty acid synthase (FAS-II) system of mycobacteria, though its precise mechanism of action has been difficult to elucidate and remains controversial. Both the enoyl-acyl carrier protein (ACP) reductase InhA (3, 26) and the β-ketoacyl-ACP synthase KasA (22, 29) have been proposed as the primary target; however, the most recent body of evidence seems to weigh in favor of InhA (19, 31). Thiolactomycin is a natural product with broad-spectrum antibiotic activity that specifically inhibits FAS-II (16). It exhibits activity against mycobacterial FAS-II (28) by its inhibition of the β-ketoacyl-ACP synthases KasA and KasB (18, 27). Its favorable physical and pharmacokinetic properties and low toxicity profile have made it an attractive candidate for a lead optimization program aimed at developing analogues with enhanced activity against M. tuberculosis (12). Triclosan is a broad-spectrum antibiotic that has been shown to inhibit InhA from M. smegmatis (21) and M. tuberculosis (25).
In order to support drug development programs aimed at the provision of novel InhA inhibitors and thiolactomycin analogues, we have, by using DNA microarrays, generated signature profiles of M. tuberculosis in response to treatment with isoniazid, thiolactomycin, and triclosan. We have compared and contrasted the response profiles to the three drugs and can distinguish between isoniazid and thiolactomycin treatment, thereby providing insight into the differences in the mechanisms of action of these two drugs. We built a predictive model, based on the expression pattern of 21 genes, which is able to perfectly classify isoniazid-, thiolactomycin-, or triclosan-treated M. tuberculosis. This can be used to evaluate novel inhibitors and will facilitate lead optimization programs by enabling the modes of action of inhibitors to be tracked, thus aiding the translation of enzyme activity to the whole cell.
MATERIALS AND METHODS
Bacterial culture and drug treatment.
Cultures were taken from frozen seed stock of M. tuberculosis strain H37Rv (NCTC 7416). Seeds were thawed, grown to late-log phase without shaking, and then diluted 1:100 into roller bottles in 100 ml of Middlebrook 7H9 medium supplemented with 0.2% (vol/vol) glycerol, 10% (vol/vol) albumin-dextrose-catalase, and 0.025% (vol/vol) Tween 80 at 37°C with constant rolling at 2 rpm. After 5 days' growth to log phase, cultures were transferred in 30-ml aliquots to 150-ml standing flasks (Nalgene) and incubated at 37°C overnight. Cultures were then treated with 10 μl of drug dissolved either in dimethyl sulfoxide for thiolactomycin and triclosan or water for isoniazid, to give final concentrations of 1× and 5× the MIC. MIC concentrations used were 0.1 μg of isoniazid per ml and 8 μg of both thiolactomycin and triclosan per ml. Control samples were treated with 10 μl of the appropriate solvent vehicle. All cultures were incubated at 37°C.
RNA isolation and preparation of labeled cDNA.
Cultures were harvested by centrifugation (1,900 × g, 15 min) after 2 or 6 h of drug treatment. Vehicle-treated control samples were harvested at time zero (t = 0) and at 2 and 6 h. Pellets were resuspended in 1 ml of TRIzol (Invitrogen), and RNA extraction was performed as previously described (6). Fluorescently labeled cDNA copies of total RNA were prepared by direct incorporation of fluorescent nucleotide analogues during a first-strand reverse transcription (RT) reaction using genome-directed priming (30). Each 25-μl labeling reaction included 2.5 μg of RNA; 1.5 μl of primer mix; 0.5 mM each of dATP, dGTP, and dCTP; 0.05 mM dTTP; 10 mM dithiothreitol (DTT); and 200 U of reverse transcriptase (Superscript II, Invitrogen) in a 1× reaction buffer provided by the enzyme manufacturer and 2 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia Biotech). The RNA and primers were preheated to 70°C for 3 min and snap-cooled on ice before adding the remaining reaction components. The RT reaction was allowed to proceed for 2 min at 25°C followed by 90 min at 42°C. Any remaining RNA was inactivated for 25 min at 37°C using RNase H (Invitrogen). All treated samples were compared to the time zero control by cohybridization. The two labeled cDNA samples to be compared were combined and purified using a QIAquick PCR purification kit (Qiagen), and eluted cDNA was dried by vacuum centrifugation.
Microarray hybridization and data analysis.
DNA microarrays consisted of 3,880 PCR-amplified open reading frame (ORF)-specific DNA fragments, representing 99% of the predicted 3,924 M. tuberculosis strain H37Rv ORFs (11), which were printed onto glass slides. Prior to hybridization, microarray slides were washed twice in isopropanol, first for 5 min and then for 10 min, before being boiled in double-distilled H2O for 5 min. Slides were then incubated at 42°C for 30 min in prehybridization buffer (1% bovine serum albumin, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]), washed twice in 0.06× SSC for 2 min, and dried by centrifugation (100 × g, 1 min). Probes were applied to the array in 40 μl of hybridization solution (5× SSC, 25% formamide, 0.5% SDS, 1× Denhardt's solution, 0.125 μg of salmon sperm DNA per ml, and 0.125 μg of Escherichia coli tRNA per ml). Samples were first denatured by heating them to 98°C for 3 min, and hybridization was carried out under a glass coverslip in a humidified slide chamber (Corning) submerged in a 42°C water bath for approximately 18 h. Coverslips were removed by incubation for 1 min in wash buffer I (2× SSC, 0.1% SDS, 1 mM DTT) prewarmed to 42°C, and slides were then washed sequentially in buffer I, buffer II (0.1× SSC, 0.1% SDS, 1 mM DTT) and twice in buffer III (0.1× SSC, 1 mM DTT) for 5 min each at room temperature. Finally, slides were dipped in 0.06× SSC for 10 s, dried by centrifugation (100 × g, 1 min), and immediately scanned using a GenePix 4000B scanner (Axon Instruments). The resulting images were analyzed using GenePix Pro 3.0 software (Axon Instruments), and data were imported into the Rosetta Resolver version 3.1 Gene Expression Data Analysis System for further analysis. An error model, calculated from 10 same-versus-same hybridizations, was applied in order to standardize log expression ratios generated from the intensity values of the treated/time zero control channels and then to quantify the significance of expression changes in each treated sample compared to the time zero control. Principal components analysis (PCA) and partial least-squares discriminant analysis (13) were performed using SIMCA-P version 10 software. Stepwise linear-discriminant analysis (15) was performed using SAS version 8 software.
Real-time quantitative RT-PCR (QRT-PCR).
cDNA was synthesized from 1 μg of RNA using Superscript II reverse transcriptase (Invitrogen) as described above but using equal concentrations of dATP, dGTP, dCTP, and dTTP (0.5 mM) as well as omitting the labeled nucleotide. After incubation at 42°C for 90 min, the cDNA was diluted to a volume of 120 μl, and 2.5 μl was used per PCR. Primers were designed using Primer Express software version 2.0 (Applied Biosystems) and the sequences shown in Table 1. PCR was performed using an Applied Biosystems 7900HT Sequence Detector System, with samples in 384-well plates. Each 10-μl reaction mixture included cDNA from 20 ng of RNA, 400 nM of each primer pair, and QuantiTect SYBR Green PCR Master Mix (Qiagen). PCR parameters used were 50°C for 2 min, 95°C for 10 min, and 33 cycles of 95°C for 15 s and 60°C for 1 min. A linear-regression line calculated from the standard curves of serially diluted M. tuberculosis H37Rv genomic DNA allowed relative transcript levels in RNA samples to be determined. Quantitative results for each cDNA were normalized to the number of sigA molecules and the significance of differential gene expression in each treated sample, relative to the nontreated, time zero control, measured using analysis of covariance (7). Analysis was performed on duplicate biological samples that were each assayed in duplicate. Chromosomal DNA contamination was measured by real-time PCR of RNA not treated with reverse transcriptase and found to be negligible.
TABLE 1.
Primers used for the QRT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| sigA | TTCGCGCCTACCTCAAACAG | GCTAGCTCGACCTCTTCCTCG |
| fabD | ACCCTGGTCTCCCAGCTCAC | AGTTCGCGTTTGGCGATACC |
| acpM | CAAGTACGGCGTCAAGATCCC | ACTTGGACTCGGCCTCAAGC |
| kasA | TGCTCATCGAGACGGAGGAG | CTACCGGCACGAACACCATC |
| kasB | CCGTGCAGAAGTACATGCCC | ACCGCTTCGATCCTGGTCTC |
| accD6 | CACACCGTTCGACGAGTTCC | TCTGCGCTTTCGGAGTTCAG |
| Rv1685c | ACTTCGGCACCAAACAGCAG | AACCGAGTTCCTCGACAGGC |
| Rv1686c | TGATGTTTGTGATCACGGCG | GAACCAGAACGCCACAATGC |
| Rv1687c | GGTCGGGCAAGACAACACTG | AGAGTTCGGCGAAGTAGCGG |
| Rv3160c | CTGGACCACCTGCATGCTTC | CCATCTCCGCGACTAGCTCC |
| Rv3161c | CAGGGTTCCCTTCACCGTTC | GCGGAATAAAGCCGAACCAC |
RESULTS
The effect of isoniazid, thiolactomycin, or triclosan treatment on M. tuberculosis gene expression was monitored using DNA microarrays. Duplicate cultures were treated with either 1× or 5× the MIC of each drug, and RNA was isolated from these as well as from the vehicle-treated control samples after either 2 or 6 h of incubation. These concentrations and time points were chosen after our own pilot studies and through consulting previously published studies (33) in order to ensure that the primary effects of the drugs and any dose responses would be captured. RNA isolated from multiple vehicle-treated cultures at time zero was pooled and used as the reference to which each sample from the later time points was compared by cohybridization. Each duplicate sample was hybridized twice, with the fluors reversed for the second batch of hybridizations. The whole-genome array was printed twice on each slide, hence giving a total of eight measurements per gene for each treatment at each time point.
Data were analyzed using Rosetta Resolver version 3.1 software, and P values representing the significance of differential expression relative to the time zero control were generated through application of a predetermined error model. Genes displaying a P value that was <0.001 were considered significantly differentially expressed relative to the time zero control. Drug-specific changes were determined by removing genes found to be differentially expressed in the vehicle control samples at the same time point with a P value of <0.05; however, genes with value changes in response to drug treatment (n-fold) of more than 1.5 times those in this vehicle control were retained as drug-induced changes.
Global gene expression patterns.
The numbers of genes, according to functional class, regulated by the drug treatments at each time point are displayed in Table 2. In general, fewer than 100 genes were regulated in response to each treatment, the exception being 5×-MIC triclosan, in which case several hundreds of genes were significantly differentially expressed. Genes encoding enzymes involved in fatty acid metabolism, oxidoreductases, and membrane proteins were among those upregulated by the 5×-MIC triclosan treatments. Genes downregulated included those encoding ribosomal proteins, fatty acid biosynthesis, and modification enzymes and proteins involved in aerobic respiration. This suggests that the 5×-MIC triclosan treatments induced many nonspecific secondary effects and genes involved in growth slowdown or cell death. In contrast, both the 1×- and 5×-MIC isoniazid or thiolactomycin treatments induced a much more specific response, including upregulation of cell wall and lipid metabolism genes.
TABLE 2.
Numbers of genes regulated in M. tuberculosis by each drug's indicated MIC, according to functional class, at 2 and 6 ha
| Functional classb | Total no. on array | 2 h
|
6 h
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1×-MIC INH
|
5×-MIC INH
|
1×-MIC TLM
|
5×-MIC TLM
|
1×-MIC TRC
|
5×-MIC TRC
|
1×-MIC INH
|
5×-MIC INH
|
1×-MIC TLM
|
5×-MIC TLM
|
1×-MIC TRC
|
5×-MIC TRC
|
||||||||||||||
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | ||
| 0: Virulence, detoxification, adaptation | 98 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 2 | 2 | 0 | 9 | 4 | 0 | 2 | 1 | 4 | 1 | 1 | 0 | 0 | 0 | 1 | 9 | 13 |
| 1: Lipid metabolism | 231 | 4 | 1 | 5 | 1 | 14 | 1 | 5 | 0 | 3 | 1 | 11 | 16 | 1 | 0 | 5 | 4 | 8 | 2 | 6 | 3 | 1 | 3 | 13 | 27 |
| 2: Information pathways | 224 | 2 | 0 | 0 | 2 | 15 | 3 | 0 | 0 | 2 | 6 | 8 | 49 | 1 | 0 | 1 | 1 | 0 | 2 | 1 | 0 | 0 | 8 | 6 | 60 |
| 3: Cell wall and cell processes | 699 | 4 | 1 | 3 | 1 | 14 | 4 | 4 | 1 | 14 | 1 | 44 | 24 | 2 | 1 | 14 | 4 | 3 | 4 | 4 | 6 | 9 | 0 | 51 | 33 |
| 5: Insertion sequences and phages | 141 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 2 | 0 | 12 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 10 | 1 |
| 6: PE/PPE | 142 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 7: Intermediary metabolism and respiration | 889 | 7 | 7 | 4 | 2 | 17 | 6 | 1 | 5 | 21 | 4 | 54 | 23 | 2 | 2 | 5 | 5 | 0 | 4 | 1 | 2 | 6 | 0 | 54 | 57 |
| 8: Unknown | 265 | 0 | 4 | 0 | 0 | 3 | 6 | 1 | 1 | 2 | 2 | 10 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 11 |
| 9: Regulatory proteins | 186 | 0 | 2 | 1 | 2 | 0 | 1 | 1 | 1 | 2 | 3 | 17 | 2 | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 25 | 4 |
| 10: Conserved hypothetical proteins | 1,005 | 7 | 10 | 6 | 2 | 14 | 9 | 1 | 5 | 11 | 7 | 42 | 42 | 2 | 3 | 9 | 4 | 2 | 1 | 4 | 1 | 7 | 0 | 87 | 56 |
| Total | 3,880 | 25 | 26 | 20 | 10 | 78 | 33 | 14 | 15 | 59 | 24 | 207 | 168 | 9 | 8 | 38 | 24 | 14 | 15 | 16 | 14 | 26 | 12 | 288 | 262 |
INH, isoniazid; TLM, thiolactomycin; TRC, triclosan; Up and Down, up- and downregulated.
According to TubercuList (http://genolist.pasteur.fr/TubercuList/).
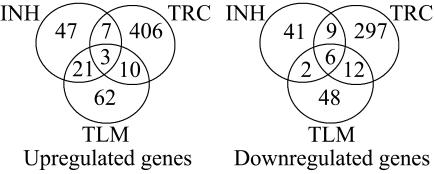
The total number of genes up- and downregulated by each drug at either dose or time point and the overlap between the drugs is displayed in Fig. 1. These Venn diagrams indicate that many genes are uniquely regulated by one drug. There is greater overlap between the genes upregulated by isoniazid and thiolactomycin than either drug with triclosan, while thiolactomycin and triclosan have the most downregulated genes in common. Genes commonly up- or downregulated by all three drug treatments are displayed in Table 3. The genes mtc28, Rv1987, and Rv3354 were induced in response to all three drugs. Two of these three genes belong to the cell wall and cell processes functional class. Both desA1 and desA2, thought to be involved in the conversion of saturated fatty acids to unsaturated fatty acids, were downregulated in response to all three drugs. In addition, Rv0823c, a possible transcriptional regulator adjacent to desA1 in the genome, was also downregulated by all three drugs. Included within the 21 genes regulated by both isoniazid and thiolactomycin (Table 3) are four of the five kas operon genes (Rv2243 to Rv2246), iniA, ahpC, efpA, and Rv1592c, all of which have been previously shown to be induced by both isoniazid or ethionamide treatment of M. tuberculosis using DNA microarrays (33).
FIG. 1.
Overlap of genes regulated by isoniazid, thiolactomycin, or triclosan treatment of M. tuberculosis. Numbers within the sectors indicate the total numbers of genes regulated uniquely or in common by either 1× or 5× MIC treatment of each drug at either 2 or 6 h (P < 0.001). INH, isoniazid; TLM, thiolactomycin; TRC, triclosan.
TABLE 3.
Genes commonly regulated by the drug treatments
| ORF | Gene name | Description | Functional classa |
|---|---|---|---|
| Induced by isoniazid, thiolactomycin, and triclosan | |||
| Rv0040c | mtc28 | Secreted proline-rich protein | 3 |
| Rv1987 | Possible chitinase | 3 | |
| Rv3354 | Conserved hypothetical protein | 10 | |
| Downregulated by isoniazid, thiolactomycin, and triclosan | |||
| Rv0352 | dnaJ1 | Probable chaperone protein | 0 |
| Rv0790c | Hypothetical protein | 8 | |
| Rv0823c | Possible transcriptional regulatory protein | 9 | |
| Rv0824c | desA1 | Probable acyl-[ACP] desaturase | 1 |
| Rv1094 | desA2 | Possible acyl-[ACP] desaturase | 1 |
| Rv2840c | Conserved hypothetical protein | 10 | |
| Induced by isoniazid and thiolactomycin | |||
| Rv0179c | lprO | Possible lipoprotein | 3 |
| Rv0207c | Conserved hypothetical protein | 10 | |
| Rv0312 | Conserved hypothetical proline- and threonine-rich protein | 10 | |
| Rv0342 | iniA | Isoniazid-inducible gene protein | 3 |
| Rv0359 | Probable conserved integral membrane protein | 3 | |
| Rv0569 | Conserved hypothetical protein | 10 | |
| Rv0951 | sucC | Probable succinyl coenzyme A synthetase (beta chain) | 7 |
| Rv1592c | Conserved hypothetical protein | 10 | |
| Rv1736c | narX | Probable nitrate reductase | 7 |
| Rv2007c | fdxA | Probable ferredoxin | 7 |
| Rv2193 | ctaE | Probable cytochrome c oxidase (subunit III) | 7 |
| Rv2243 | fabD | Malonyl coenzyme A-acyl carrier protein transacylase | 1 |
| Rv2244 | acpM | Meromycolate extension acyl carrier protein | 1 |
| Rv2245 | kasA | β-Ketoacyl-[ACP] synthase 1 | 1 |
| Rv2246 | kasB | β-Ketoacyl-[ACP] synthase 2 | 1 |
| Rv2428 | ahpC | Alkyl hydroperoxide reductase C | 0 |
| Rv2721c | Possible conserved transmembrane alanine- and glycine-rich protein | 3 | |
| Rv2846c | efpA | Possible integral membrane efflux protein | 3 |
| Rv3134c | Conserved hypothetical protein | 10 | |
| Rv3456c | rplQ | Probable 50S ribosomal protein L17 | 2 |
| Rv3524 | Probable conserved membrane protein | 3 |
Functional classes are listed in Table 1.
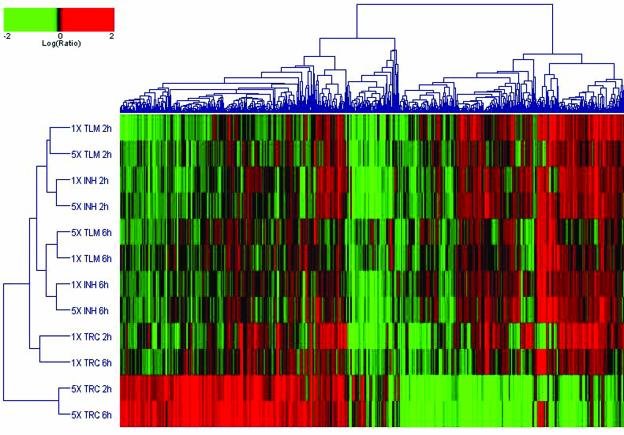
Two-dimensional clustering of expression profiles was performed with Resolver using the set of 877 genes found to be significantly regulated by any of the drug treatments (Fig. 2). This cluster analysis groups expression profiles together on the basis of their similarity across the subset of genes, and it simultaneously organizes the genes by similarities in their expression pattern. The length and subdivision of the dendrogram branches indicate the extent of similarity between the treatment-induced expression profiles (y axis) or the individual gene expression patterns (x axis). The drug expression profiles clustered into two main groups. The 5×-MIC triclosan treatments at both the 2- and 6-h time points clustered together in one outlying group and displayed a very different expression profile from that of the other treatments. In the other group, the 1×-MIC triclosan treatments clustered together and did so separately from the thiolactomycin and isoniazid profiles, which were more closely related to each other and clustered according to time.
FIG. 2.
Two-dimensional cluster analysis of the drug-treated expression profiles. Two-dimensional agglomerative clustering was performed on the 877 genes significantly regulated in response to any of the drug treatments (P < 0.001). The individual genes are represented on the x axis and the different samples are indicated on the y axis. Red, upregulation; green, downregulation; black, no change relative to the time zero control. INH, isoniazid; TLM, thiolactomycin; TRC, triclosan.
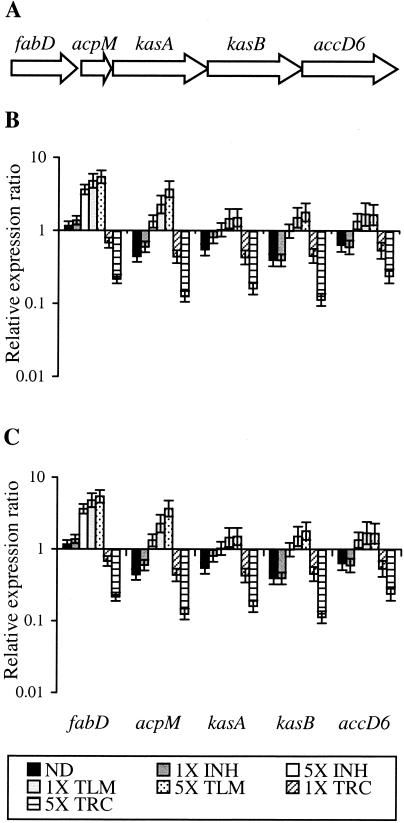
Effects of isoniazid, thiolactomycin, and triclosan treatment on the kas operon.
The most highly induced genes in response to both isoniazid and thiolactomycin were members of the kas operon (Rv2243-Rv2247). Both drugs have previously been shown to upregulate this operon, which contains five FAS-II components, including kasA and kasB (29, 33). Our microarray data confirmed these findings and showed that this operon is induced by the 5×-MIC isoniazid treatments and both the 1×- and 5×-MIC thiolactomycin treatments (Table 4). The highest induction of the kas operon in response to isoniazid treatment was observed with the 5×-MIC dose at the 6-h time point, while the maximum induction measured in response to thiolactomycin treatment was with the 1×-MIC dose at 2 h. The differences in the kinetics of the response of this operon to either isoniazid or thiolactomycin treatment may reflect differences in the mechanism of action of these two drugs. In contrast to the induction observed as a result of isoniazid or thiolactomycin treatment, expression of the kas operon was downregulated by the 5×-MIC triclosan treatments (Table 4), with the greatest degree of downregulation observed at the 6-h time point. The effects of the 1×-MIC isoniazid and 1×-MIC triclosan treatments on this operon were not significant.
TABLE 4.
Differential response of the kas operon to isoniazid, thiolactomycin, or triclosan treatment
| Treatmenta and time |
fabD
|
acpM
|
kasA
|
kasB
|
accD6
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratiob | P value | Ratio | P value | Ratio | P value | Ratio | P value | Ratio | P value | |
| 5×-MIC INH, 2 h | 1.53 | 3.06 × 10−4 | 3.15 | <1.00 × 10−46 | 2.81 | 2.78 × 10−29 | 2.78 | 1.49 × 10−28 | NS | |
| 5×-MIC INH, 6 h | 2.27 | <1.00 × 10−46 | 4.53 | <1.00 × 10−46 | 4.40 | 1.87 × 10−31 | 3.54 | <1.00 × 10−46 | NS | |
| 1×-MIC TLM, 2 h | 2.26 | 1.54 × 10−4 | 4.30 | <1.00 × 10−46 | 3.79 | 4.77 × 10−15 | 3.42 | 1.09 × 10−18 | 1.69 | 1.12 × 10−11 |
| 5×-MIC TLM, 2 h | NS | 3.70 | <1.00 × 10−46 | 2.94 | 7.92 × 10−20 | 2.08 | 1.43 × 10−6 | 1.41 | 8.06 × 10−5 | |
| 1×-MIC TLM, 6 h | 1.88 | 9.48 × 10−6 | 3.01 | <1.00 × 10−46 | 2.87 | 5.33 × 10−39 | 2.10 | 8.86 × 10−10 | 1.41 | 8.36 × 10−4 |
| 5×-MIC TLM, 6 h | 1.79 | 5.01 × 10−6 | 4.00 | <1.00 × 10−46 | 2.93 | 4.49 × 10−37 | 2.41 | 1.61 × 10−14 | NS | |
| 5×-MIC TRC, 2 h | NS | 0.24 | 3.30 × 10−24 | 0.30 | 8.29 × 10−15 | 0.36 | 6.27 × 10−9 | NS | ||
| 5×-MIC TRC, 6 h | NS | 0.13 | <1.00 × 10−46 | 0.20 | <1.00 × 10−46 | 0.34 | 5.37 × 10−25 | NS | ||
INH, isoniazid; TLM, thiolactomycin; TRC, triclosan.
Average expression ratio (drug treated/t = 0 control) calculated from a total of four hybridizations of two biological replicates. NS, not significant (P < 0.001).
Using an independent RNA sample set, we then went on to perform QRT-PCR analysis of the kas operon genes in response to the drug treatments (Fig. 3). This confirmed the induction of the kas operon in response to the 5×-MIC isoniazid treatment and both thiolactomycin doses but also confirmed downregulation in response to the 5×-MIC triclosan treatment and hence validated the microarray data in an independent sample set. The QRT-PCR data showed more clearly that induction in response to the 5×-MIC isoniazid treatment was greater at the 6-h time point, while thiolactomycin treatment gave a more immediate high-level induction at 2 h. The QRT-PCR data also confirmed that both the 1×-MIC isoniazid and 1×-MIC triclosan treatments had little effect on the expression of the kas operon compared to the vehicle control.
FIG. 3.
Response of the kas operon to isoniazid, thiolactomycin, or triclosan treatment as measured by QRT-PCR. (A) Schematic representation of the kas operon in the M. tuberculosis H37Rv genome. Ratio between the number of cDNA copies detected in each sample relative to the time zero control by QRT-PCR at 2 h (B) and 6 h (C) is represented. Each value is the average of two biological replicates, each analyzed in duplicate. INH, isoniazid; TLM, thiolactomycin; TRC, triclosan.
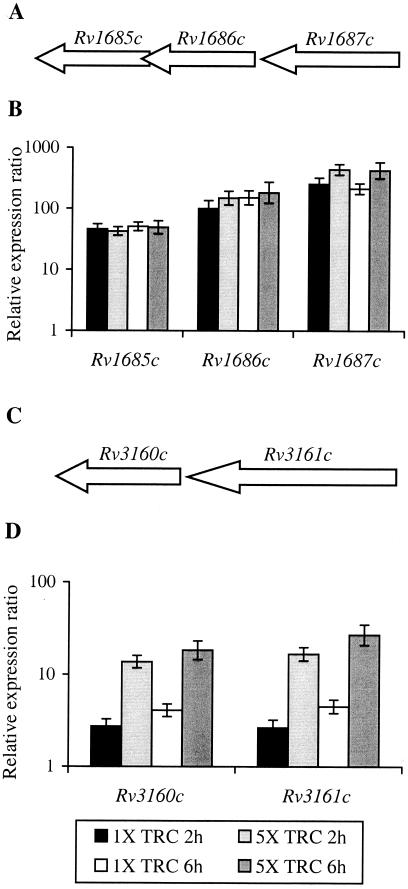
Triclosan-induced genes.
Eight genes were induced in response to all triclosan treatments (Table 5). Of these eight genes, only mmpL6 was also regulated by either isoniazid or thiolactomycin, being induced by the 1×-MIC thiolactomycin treatment at 2 h. Two groups of genes, probably forming operons, were highly induced in response to all triclosan treatments. Both sets of genes, Rv1685c to Rv1687c and Rv3160c to Rv3161c, were most highly induced in response to the 5×-MIC triclosan treatments. QRT-PCR analysis of these genes in an independent RNA sample set confirmed their induction in response to triclosan treatment (Fig. 4) and their lack of regulation in response to either isoniazid or thiolactomycin or in the vehicle control (data not shown) and hence validated the microarray findings. Induction levels as measured by QRT-PCR were generally higher than those observed using DNA microarrays, probably due to the greater sensitivity and dynamic range achievable using quantitative PCR techniques rather than two-color microarrays. This was most apparent with genes Rv1685c to Rv1687c, for which induction levels measured using QRT-PCR were up to 38 times higher than those observed using the microarray.
TABLE 5.
Genes induced in M. tuberculosis by all triclosan (TRC) treatments
| ORF | Gene name | Description | Functional classa | MIC and exposure time
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1×-MIC TRC, 2 h
|
5×-MIC TRC, 2 h
|
1×-MIC TRC, 6 h
|
5×-MIC TRC, 6 h
|
||||||||
| Ratiob | P value | Ratio | P value | Ratio | P value | Ratio | P value | ||||
| Rv0077c | Probable oxidoreductase | 7 | 1.88 | 1.05 × 10−6 | 5.46 | 3.33 × 10−8 | 1.60 | 1.87 × 10−11 | 3.57 | 7.01 × 10−45 | |
| Rv0711 | atsA | Possible arylsulfatase | 7 | 2.64 | 4.48 × 10−17 | 3.16 | <1.00 × 10−46 | 1.65 | 4.40 × 10−7 | 1.92 | 1.86 × 10−29 |
| Rv1557 | mmpL6 | Probable conserved transmembrane transport protein | 3 | 2.16 | 5.31 × 10−16 | 1.67 | 2.71 × 10−12 | 1.41 | 9.25 × 10−8 | 1.66 | 2.59 × 10−19 |
| Rv1685c | Conserved hypothetical protein | 10 | 17.89 | <1.00 × 10−46 | 21.20 | 3.06 × 10−26 | 13.15 | <1.00 × 10−46 | 21.17 | <1.00 × 10−46 | |
| Rv1686c | Probable conserved integral membrane protein, ABC transporter | 3 | 14.99 | <1.00 × 10−46 | 16.58 | <1.00 × 10−46 | 11.02 | <1.00 × 10−46 | 21.30 | <1.00 × 10−46 | |
| Rv1687c | Probable conserved ATP-binding protein, ABC transporter | 3 | 6.95 | 3.49 × 10−26 | 12.95 | 1.26 × 10−17 | 5.06 | <1.00 × 10−46 | 11.16 | <1.00 × 10−46 | |
| Rv3160c | Possible transcriptional regulatory protein (probably TetR family) | 9 | 2.40 | 4.62 × 10−6 | 9.51 | 2.34 × 10−18 | 2.01 | 3.35 × 10−9 | 8.63 | <1.00 × 10−46 | |
| Rv3161c | Possible dioxygenase | 7 | 4.77 | 1.03 × 10−39 | 18.92 | <1.00 × 10−46 | 4.40 | <1.00 × 10−46 | 18.42 | <1.00 × 10−46 | |
Functional classes are listed in Table 1.
Average expression ratio (drug treated/t = 0 control) calculated from a total of four hybridizations of two biological replicates.
FIG. 4.
Genes induced by triclosan treatment of M. tuberculosis as measured by QRT-PCR. Organization of Rv1685c to Rv1687c (A) and Rv3160c to Rv3161c (C) in the M. tuberculosis H37Rv genome. Ratio between the number of cDNA copies detected in each sample relative to the time zero control by QRT-PCR for Rv1685c to Rv1687c (B) and Rv3160c to Rv3161c (D). Each value is the average of two biological replicates, each analyzed in duplicate. TRC, triclosan.
Using gene expression data to classify drug treatments.
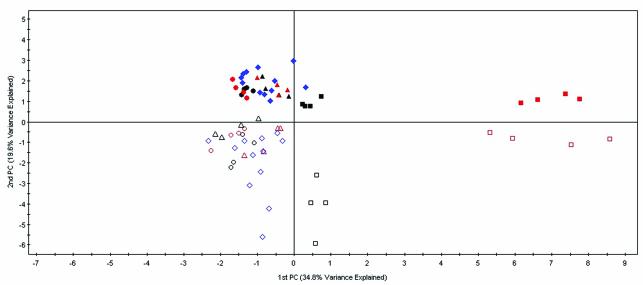
With the aim of obtaining a model that would be able to classify M. tuberculosis treated with either isoniazid, thiolactomycin, triclosan, or with no drug and that would have utility in our drug development program, we went on to perform further statistical analysis of the microarray data. PCA was first performed to identify the main sources of variability between the three drug treatments and treatment controls (Fig. 5). PCA is a technique that enables a high-dimensional data set to be represented by two or three variables (known as principal components). The principal components are estimated so that they represent the main structural features and variability in the data. Each principal component is made up of a linear combination of all the gene intensities. The higher the influence a gene has on a principal component, the larger is its effect on the position of a point on the score plot. Each point on the score plot shown in Fig. 5 represents a single hybridization. The largest source of variance (34.8%) is shown on the x axis and was accounted for by separating the 5×-MIC dose of triclosan from the other groups. The next largest source of variance (19.6%) was time, displayed on the y axis, with the 6-h treatments clustering above the axis and the 2-h treatments below. This correlates with the two-dimensional cluster analysis, in which the isoniazid and thiolactomycin profiles clustered according to time (Fig. 2). Within these groups, the isoniazid and thiolactomycin treatments tend to group together, while the 1×-MIC triclosan treatments are grouped together as outliers.
FIG. 5.
PCA of isoniazid, thiolactomycin, triclosan, and control expression profiles. The largest source of variance is explained on the x axis and the second largest on the y axis. Each hybridization is represented by a single point. Isoniazid treatments, circles; thiolactomycin treatments, triangles; triclosan treatments, squares; vehicle control treatments, diamonds. Two-hour treatments, open shapes; 6-h treatments, closed shapes. 1× MIC, black; 5× MIC, red; vehicle control, blue.
The 5×-MIC doses of each drug at the 6-h time point were found to be the most clearly separated from each other and from the vehicle controls. These data sets were therefore analyzed using partial least-squares discriminant analysis in order to score genes as to their relative importance for discriminating between the four groups—isoniazid, thiolactomycin, triclosan, or control treatment of M. tuberculosis, at this dose and time point. Interestingly, the top 35 genes from the partial least-squares analysis included all five members of the kas operon, with acpM and kasA being the two most important genes for separating the groups. Also included were Rv1685c to Rv1687c, Rv3160c to Rv3161c, and iniA and iniB, which have previously been shown to be induced by isoniazid treatment of M. tuberculosis (1, 33). Here we found the 5×-MIC isoniazid treatment at 6 h to induce both iniA and iniB and found the 1×-MIC thiolactomycin treatment at 6 h to induce iniA.
Stepwise linear-discriminant analysis of the top 500 ranking genes from the partial least-squares analysis was then performed in order to generate a mathematical function capable of discriminating between the four groups for the 5×-MIC treatment at 6 h. Stepwise linear-discriminant analysis builds a linear model by selecting a small subset of genes that best discriminate among the groups. The model was built using the data obtained from the original hybridizations for each of the four treatment groups—isoniazid, thiolactomycin, triclosan, or treatment control—giving a total of 24 expression profiles. A function involving 21 genes (Table 6) was found to perfectly classify each of the expression profiles into one of the four groups. The equation for the discriminant function is shown in Table 6. All 21 genes were required for perfect classification, with the first three genes accounting for 85% of the variance between the four groups and Rv1686c and acpM being the two most important discriminant genes. The experiment was then repeated in the same format to generate a set of independent data in order to test the model. A total of four hybridizations of two biological replicates was performed for each of the four treatments, giving a total of 16 expression profiles. The test data were classified into one of the four groups based on a probability score defining how closely each profile fitted the model for that group. All 16 expression profiles classified correctly into their respective groups with a probability of >0.99, thus providing independent validation of the model.
TABLE 6.
The genes and discriminant function required for classification of isoniazid (INH), thiolactomycin (TLM), triclosan (TRC), or control groups
| ORF | Gene name | Description | Avg squared canonical correlationa | INH [D2INH (x)]b | TLM [D2TLM (x)] | TRC [D2TRC (x)] | Control [D2Control (x)] |
|---|---|---|---|---|---|---|---|
| Constant | −14,987,826 | −167,097,220 | −79,450,033 | −5,513,637 | |||
| Rv1686c | Probable conserved integral membrane protein ABC transporter | 0.3325 | −19,099,766 | −100,023,236 | −36,942,559 | −19,032,909 | |
| Rv2244 | acpM | Meromycolate extension acyl carrier protein | 0.6111 | 110,774,262 | 408,156,465 | 243,639,840 | 72,779,372 |
| Rv0677c | mmpS5 | Possible conserved membrane protein | 0.8533 | 121,600,815 | 512,108,617 | 316,180,097 | 94,776,642 |
| Rv2190c | Conserved hypothetical protein | 0.8904 | −68,508,665 | −286,328,959 | −181,659,649 | −52,963,842 | |
| Rv3250c | rubB | Probable rubredoxin | 0.8954 | −10,150,989 | −44,990,430 | −22,707,705 | −8,353,423 |
| Rv2276 | cyp121 | Cytochrome P450 121 | 0.9040 | −36,833,310 | −220,327,670 | −143,773,133 | −43,835,221 |
| Rv3049c | Probable monooxygenase | 0.9491 | −114,909,397 | −452,847,559 | −265,224,702 | −82,194,446 | |
| Rv0208c | Hypothetical methyltransferase | 0.9762 | −178,128,313 | −821,139,430 | −524,976,195 | −155,439,247 | |
| Rv2745c | Possible transcriptional regulatory protein | 0.9817 | −77,131,242 | −323,842,646 | −200,033,735 | −59,890,920 | |
| Rv2253 | Possible secreted unknown protein | 0.9834 | −49,162,506 | −201,531,571 | −124,495,122 | −37,049,967 | |
| Rv1072 | Probable conserved transmembrane protein | 0.9843 | 64,061,474 | 246,735,357 | 152,824,150 | 44,629,671 | |
| Rv0298 | Hypothetical protein | 0.9865 | 47,211,509 | 172,163,109 | 103,076,691 | 30,611,076 | |
| Rv3341 | metA | Probable homoserine O-acetyltransferase | 0.9867 | 1,400,025 | −26,153,983 | −29,413,890 | −6,482,707 |
| Rv0001 | dnaA | Chromosomal replication initiator protein | 0.9932 | −23,069,217 | −86,660,290 | −56,244,876 | −15,604,890 |
| Rv3457c | rpoA | Probable DNA-directed RNA polymerase (α chain) | 0.9942 | −82,964,894 | −232,795,471 | −118,734,295 | −37,450,414 |
| Rv1737c | narK2 | Possible nitrate/nitrite transporter | 0.9946 | −22,730,982 | −62,077,538 | −26,268,313 | −9,791,891 |
| Rv1871c | Conserved hypothetical protein | 0.9967 | 16,814,613 | 46,910,792 | 21,021,772 | 7,485,856 | |
| Rv3404c | Conserved hypothetical protein | 0.9971 | 46,788,129 | 124,430,283 | 61,274,979 | 19,520,565 | |
| Rv3486 | Conserved hypothetical protein | 0.9972 | −63,601,803 | −175,065,603 | −83,869,339 | −27,853,590 | |
| Rv3310 | Possible acid phosphatase | 0.9998 | 35,921,002 | 97,302,396 | 47,325,500 | 15,382,806 | |
| Rv0283 | Possible conserved membrane protein | 1.0000 | 22,910,896 | 62,309,712 | 30,137,464 | 9,865,584 |
Cumulative canonical correlation showing the increasing amount of variance accounted for by each gene in the discriminant model.
To classify an observation (x) into one of the groups, the discriminant score [D2y (x)] is calculated for each group (y) using the above functions. These scores are inversely related to the distance that observation is from the middle of each group. The observation belongs to the group whose function gives the highest discriminant score. The probability of an observation (x) belonging to a group (y) is calculated as eDy2(x)/[eDControl2(x)+eDINH2(x)+eDTLM2(x)+eDTRC2(x)].
DISCUSSION
Genomic technologies offer the promise of accelerating many phases of drug discovery and development from the improved selection of targets through aiding clinical trials. DNA microarrays are currently one of the most powerful of these tools, enabling the measurement of gene expression profiles on a genome-wide scale. We used microarrays to examine the response of M. tuberculosis to treatment with three mycolic acid biosynthesis inhibitors—isoniazid, thiolactomycin and triclosan, whose modes of action have been studied previously. This has generated signature expression profiles of each drug, giving insight into the effects of the drugs on the metabolism of M. tuberculosis and providing a reference to which to compare novel inhibitors from high-throughput screens or lead optimization programs.
As thiolactomycin and triclosan both have well-defined targets, KasA/KasB and InhA, respectively, they have previously been used as tools to study the molecular mechanism of isoniazid and the relative contributions of InhA and KasA in an effort to address the controversy surrounding the primary target of isoniazid (29). Our clustering and PCA analyses have shown isoniazid expression profiles to more closely resemble thiolactomycin than triclosan profiles. However, the isoniazid and thiolactomycin profiles were distinct from one another, and more genes were regulated uniquely in response to each drug than were in common with the other treatments. There were also differences in the regulation of the kas operon in response to isoniazid and thiolactomycin. Although the kas operon genes were induced by the 5×-MIC isoniazid treatment to levels similar to those observed with thiolactomycin treatment, the kinetics of the response appeared to be slower in response to isoniazid, perhaps reflecting an indirect effect. In addition, the 1×-MIC isoniazid dose did not significantly induce the kas operon, more closely resembling the 1×-MIC triclosan treatment, which also had no effect on the expression of these genes beyond that of the treatment control. Based on these observations, it seems likely that the similarities that exist in the response profiles toward isoniazid and thiolactomycin are due to both drugs inhibiting the same pathway but that differences are seen due to the drugs inhibiting different primary targets. The induction of the kas operon in response to isoniazid treatment may therefore be a secondary effect of the drug, reflected in the slower kinetics of induction compared to thiolactomycin and the absence of induction in response to 1×-MIC levels of drug. In supporting different primary targets for isoniazid and thiolactomycin, these findings would therefore support InhA rather than KasA as the primary target of isoniazid. In their study, using a luciferase reporter strain, Slayden and colleagues showed induction of the kas operon in response to isoniazid even at concentrations below the MIC (29). This difference may be due to the greater sensitivity of the luciferase assay compared to microarray analysis or the fact that cultures were incubated with the drug for 24 h prior to measuring expression levels. In a previous study using DNA microarrays, isoniazid was found to induce the kas operon after 1 h of exposure, but this was in response to concentrations equivalent to either 2× or 10× the MIC that was used in this study (33). In agreement with previous studies (33), we have not observed induction of inhA in response to isoniazid treatment. The lack of inhA induction following isoniazid treatment may represent one of the properties of an ideal target whose inhibition leads to a bactericidal event (31). The failure of a drug to induce its own target gene would preclude transient resistance and increase its effectiveness.
A recent study into the membranotropic effects of triclosan has suggested that the antibacterial effects of the drug are mediated, at least in part, through intercalation into the membrane, resulting in destabilizing structures and interfering with normal membrane-dependent processes (32). The results presented here, showing upregulation of many genes encoding membrane proteins in response to triclosan treatment, would support this mechanism in M. tuberculosis. The difference between the triclosan and the isoniazid and thiolactomycin response profiles may therefore be due to inefficient entry of triclosan into the cell, leading to low intracellular concentration of drug and hence poor inhibition of the target enzyme. In this case, response profiles would not reflect the effects of inhibition of the target enzyme but would instead reflect other nonspecific effects such as those seen for triclosan in the present study.
Further insight into the apparent failure of triclosan to effectively inhibit its target enzyme is given by the two groups of genes that were most highly induced in response to triclosan treatment. Rv1685c encodes a conserved hypothetical protein of unknown function but which has some similarity to possible transcription regulators of M. tuberculosis, including Rv3160c and a putative transcriptional regulator from Streptomyces coelicolor. Rv1686c and Rv1687c encode an integral membrane protein and an ATP-binding protein of an ABC transport system, respectively. Bioinformatic analysis of the protein sequence of this ABC transporter has found it to cluster together with other M. tuberculosis transporters similar to known antibiotic resistance systems (8). As this transporter is induced in response to triclosan treatment, it is possible that it is involved in antibiotic resistance in M. tuberculosis through provision of an efflux mechanism to export the drug from the cell. Rv3160c encodes a possible transcriptional regulator of the TetR-AcrR family, and Rv3161c encodes a possible dioxygenase with similarity to several bacterial aromatic dioxygenases involved in the degradation of arenes. As triclosan consists of a diphenyl ether structure, it is possible that induction of this enzyme could serve to degrade—and hence detoxify—triclosan. Therefore, the apparent lack of a triclosan effect on its target may be due to the induction of a triclosan efflux pump and detoxification system with resultant low intracellular concentration of triclosan. This would explain the discrepancy between the 50% inhibitory concentration for triclosan against InhA in vitro, which is lower than that for isoniazid, and the poor whole-cell activity reflected by its high MIC.
Generation of drug treatment response profiles such as those demonstrated in this study has several applications within the drug discovery process (4, 10). Signature profiles of drugs of known modes of action can be used to predict the modes of action of novel inhibitors identified through whole-cell screens. Promoters induced upon inhibition of a target pathway may be used to create reporter strains for use in an in vivo whole-cell screening approach to identify compounds that specifically inhibit the desired pathway. A further application would be the use of response profiles to confirm that a particular compound provides selective inhibition of the intended target (10, 20). A common problem encountered in the development of antimycobacterials is translating activity against an enzyme in vitro into whole-cell activity against M. tuberculosis. Triclosan is an example of a drug that has good enzyme activity but poor whole-cell activity. Microarray profiling would aid the identification of such compounds. For example, induction of the putative ABC transporter seen in response to triclosan could serve as a marker of poor cell wall penetration and could be used to identify compounds likely to have poor whole-cell activity against M. tuberculosis. The predictive model described here provides a mechanism for classifying M. tuberculosis treated with isoniazid, thiolactomycin, triclosan, or no drug. Any expression profile not fitting the model for one of these four groups would be classified as “other.” In its present form, the model is a valuable tool for confirming that thiolactomycin analogues or novel InhA inhibitors are inhibiting the correct target within the whole cell and will have significant impact on our drug discovery programs in these areas. Future generation of M. tuberculosis response profiles to drugs that target different pathways as well as further refinement of the model will extend its use to the classification of a wider range of inhibitors.
Acknowledgments
We thank Kevin Kershner for printing the microarray slides and Kay Tatsuoka for generation of the error model. We also thank George Fletcher and Tim Fulton for providing Resolver support.
REFERENCES
- 1.Alland, D., I. Kramnik, T. R. Weisbrod, L. Otsubo, R. Cerny, L. P. J. W. Miller, and B. R. Bloom. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL)—the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bammert, G., and J. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, A., E. Dubnau, A. Quemard, A. S. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. W. de Lisle, and W. R. Jacobs. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 4.Barry, C. E., M. Wilson, R. Lee, and G. K. Schoolnik. 2000. DNA microarrays and combinatorial chemical libraries: tools for the drug discovery pipeline. Int. J. Tuberc. Lung Dis. 4:S189-S193. [PubMed] [Google Scholar]
- 5.Betts, J. C. 2002. Transcriptomics and proteomics: tools for the identification of novel drug targets and vaccine candidates for tuberculosis. IUBMB Life 53:239-242. [DOI] [PubMed] [Google Scholar]
- 6.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 7.Bond, B., D. Virley, N. Cairns, A. Hunter, G. Moore, S. Moss, A. Mudge, F. Walsh, E. Jazin, and P. Preece. 2002. The quantification of gene expression in an animal model of brain ischaemia using TaqMan real-time RT-PCR. Mol. Brain Res. 106:101-116. [DOI] [PubMed] [Google Scholar]
- 8.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24:449-467. [DOI] [PubMed] [Google Scholar]
- 9.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 10.Chan, P., R. Macarron, D. Payne, M. Zalacain, and D. Holmes. 2002. Novel antibacterials: a genomics approach to drug discovery. Curr. Drug Targets Infect. Disord. 2:291-308. [DOI] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. G. S. Harris, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Conner, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osbourne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Douglas, J. D., S. J. Senior, C. Morehouse, B. Phetsukiri, I. B. Campbell, G. S. Besra, and D. E. Minnikin. 2002. Analogues of thiolactomycin: potential drugs with enhanced anti-mycobacterial activity. Microbiology 148:3101-3109. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, L., E. Johansson, N. Kettaneh-Wold, and S. Wold. 1999. Introduction to multi- and megavariate data analysis using projection methods (PCA and PLS). Umetrics.
- 14.Gmuender, H., K. Kuratli, K. Di Padova, C. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastie, T., R. Tibshirani, and J. Friedman. 2001. The elements of statistical learning—data mining, inference, and prediction. Springer-Verlag, New York, N.Y.
- 16.Hayashi, T., O. Yamamoto, H. Sasaki, H. Okazaki, and A. Kawaguchi. 1984. Inhibition of fatty acid synthesis by the antibiotic thiolactomycin. J. Antibiot. 37:1456-1461. [DOI] [PubMed] [Google Scholar]
- 17.Heath, R. J., S. W. White, and C. O. Rock. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58:695-703. [DOI] [PubMed] [Google Scholar]
- 18.Kremer, L., J. D. Douglas, A. R. Baulard, C. Morehouse, M. R. Guy, D. Alland, L. G. Dover, J. H. Lakey, W. R. Jacobs, Jr., P. J. Brennan, D. E. Minnikin, and G. S. Besra. 2000. Thiolactomycin and related analogues as novel anti-mycobacterial agents targeting KasA and KasB condensing enzymes in Mycobacterium tuberculosis. J. Biol. Chem. 275:16857-16864. [DOI] [PubMed] [Google Scholar]
- 19.Larsen, M. H., C. Vilcheze, L. Kremer, G. S. Besra, L. Parsons, M. Salfinger, L. Heifets, M. H. Hazbon, D. Alland, J. C. Sacchettini, and W. R. Jacobs. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 46:453-466. [DOI] [PubMed] [Google Scholar]
- 20.Marton, M., J. DeRisi, H. Bennett, V. Iyer, M. Meyer, C. Roberts, R. Stoughton, J. Burchard, D. Slade, H. Dai, D. Bassett, L. Hartwell, P. Brown, and S. Friend. 1998. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nature 4:1293-1301. [DOI] [PubMed] [Google Scholar]
- 21.McMurry, L., P. McDermott, and S. Levy. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob. Agents Chemother. 43:711-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mdluli, K., R. A. Slayden, Y. Q. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. M. J. Crane, and C. E. Barry. 1998. Inhibition of a Mycobacterium tuberculosis β-ketoacyl ACP synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 23.Ng, W., K. Kazmierczak, G. Robertson, R. Gilmour, and M. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien, R. J., and P. P. Nunn. 2001. The need for new drugs against tuberculosis—obstacles, opportunities, and next steps. Am. J. Respir. Crit. Care Med. 163:1055-1058. [DOI] [PubMed] [Google Scholar]
- 25.Parikh, S. L., G. P. Xiao, and P. J. Tonge. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39:7645-7650. [DOI] [PubMed] [Google Scholar]
- 26.Quemard, A., J. C. Sacchettini, A. Dessen, C. Vilcheze, R. Bittman, W. R. Jacobs, and J. S. Blanchard. 1995. Enzymic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34:8235-8241. [DOI] [PubMed] [Google Scholar]
- 27.Schaeffer, M. L., G. Agnihotri, C. Volker, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029-47037. [DOI] [PubMed] [Google Scholar]
- 28.Slayden, R. A., R. E. Lee, J. W. Armour, A. M. Cooper, I. M. Orme, P. J. Brennan, and G. S. Besra. 1996. Antimycobacterial action of thiolactomycin: an inhibitor of fatty acid and mycolic acid synthesis. Antimicrob. Agents Chemother. 40:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slayden, R. A., R. E. Lee, and C. E. Barry. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514-525. [DOI] [PubMed] [Google Scholar]
- 30.Talaat, A. M., P. Hunter, and S. A. Johnston. 2000. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 18:679-682. [DOI] [PubMed] [Google Scholar]
- 31.Vilcheze, C., H. R. Morbidoni, T. R. Weisbrod, H. Iwamoto, M. Kuo, J. C. Sacchettini, and W. R. Jacobs. 2000. Inactivation of the inhA-encoded fatty acid synthase II (FASII) enoyl-acyl carrier protein reductase induces accumulation of the FASI end products and cell lysis of Mycobacterium smegmatis. J. Bacteriol. 182:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villalain, J., C. Mateo, F. Aranda, S. Shapiro, and V. Micol. 2001. Membranotropic effects of the antibacterial agent triclosan. Arch. Biochem. Biophys. 390:128-136. [DOI] [PubMed] [Google Scholar]
- 33.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]