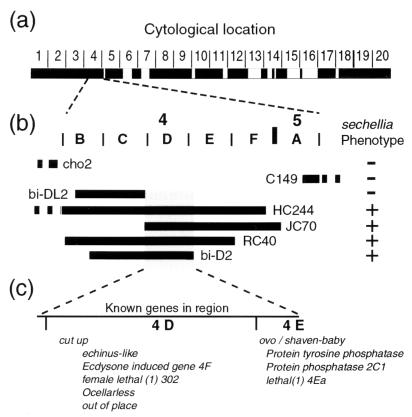
Figure 2.
Localization of the evolved gene by failure of complementation of X chromosome deficiencies. (a) The cytological regions covered by deficiencies that produced viable larvae when crossed to D. sechellia males are shown as black boxes. Approximately 75% of the chromosome was screened successfully with deficiencies. In the original screen, only Df(1)JC70 produced larvae with the D. sechellia hair pattern in the expected ratio (n = 57).[Some of the crosses yielded single larvae with a D. sechellia hair pattern. These larvae are likely the result of meiotic nondisjunction in the female parent generating nullo X eggs fertilized by X-bearing D. sechellia sperm, which in turn generated XO embryos displaying the D. sechellia hair pattern. Nondisjunction is elevated in stocks carrying balancer chromosomes and stocks with XXY females (37)]. (b) Further localization to the 4C15-E1 cytological region was performed with overlapping deficiencies. Regions deleted by deficiencies are indicated by bold horizontal lines with the name of the deficiency next to the line. The continuation of a deficiency outside this region is shown by a dashed line. Deficiencies producing larvae with a D. sechellia hair pattern in the expected frequencies when crossed to D. sechellia males are indicated with a plus sign. Deficiencies producing only larvae with a hair pattern typical of D. melanogaster are indicated by a minus sign. The distal limit of the evolved gene is defined by the right breakpoint of Df(1)bi-DL2 (4C15-D1) and the left breakpoint of Df(1) JC70 (4C15–16), and the proximal limit is defined by the right breakpoint of Df(1)bi-D2 (4D7-E1). (c) Genes known to exist within this region are listed in their approximate cytological location. The genes cut up and ovo/svb have been localized previously to the small regions, 4D1–3 and 4E1, respectively, and the three genes Protein tyrosine phosphatase 4E, Protein phosphatase 2C1, and lethal(1)4Ea have been localized previously to 4E1–2. The remaining genes shown have been localized to large regions that include 4D-E1 (see http://flybase.bio.indiana.edu for details).