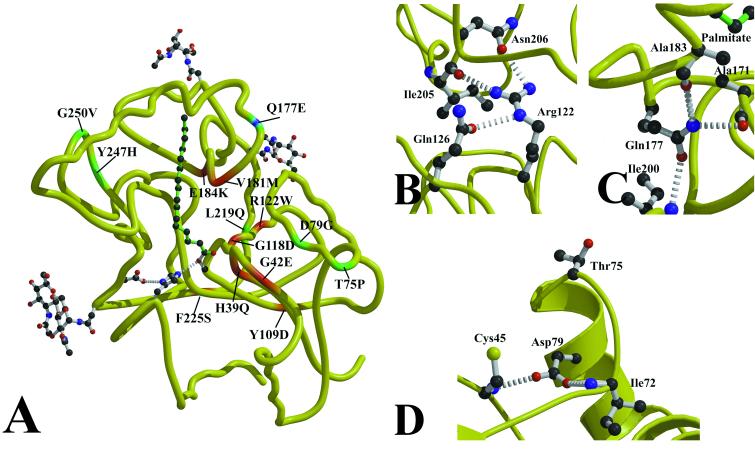
Figure 5.
NCL mutations in PPT1. (A) Sites of clinical NCL mutations in PPT1 are mapped onto the peptide backbone. INCL mutations are displayed in red, a mutation causing LINCL symptoms is in blue, and JNCL mutations are in green. (B) The most common INCL mutation (Arg122Trp) leads to the loss of three hydrogen bonds and a steric and polarity mismatch with the surrounding residues, resulting in misfolded protein. (C) The Gln177Glu mutation is predicted to cause the loss of hydrogen bonds to Ala171 and Ala183, two residues that contact palmitate. This mutation results in a less severe phenotype that is clinically indistinguishable from classical LINCL. (D) Two mutations on α1 lead to JNCL. Trp75Pro may alter the beginning of α1 due to the conformational restraints on Pro, and Asp79Glu loses hydrogen bonds to Cys45 and Ile72.