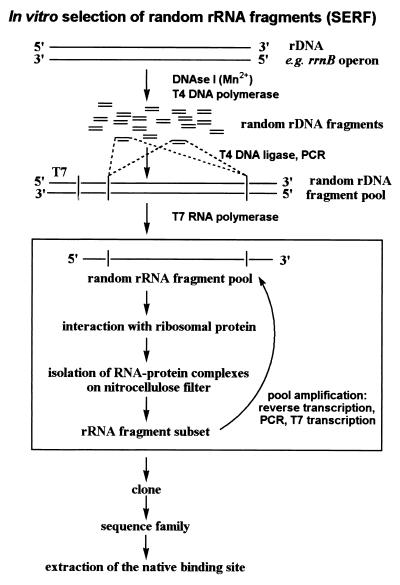
Figure 1.
Outline of the experimental strategy for SERF: rDNA is digested with DNase I in a manganese-dependent manner producing random rDNA fragments. The fragments are blunt-ended and ligated into a vector, thereby introducing constant primer regions. An rDNA fragment pool is amplified by PCR, and the pool of random rRNA fragments is prepared by T7 in vitro transcription. A certain size distribution of the fragments results from the area of RNA cut out from a denaturing acrylamide gel during purification of the RNA. The rRNA–protein complexes are formed and collected on nitrocellulose filters. Bound rRNA is recovered from the filter, reverse-transcribed to cDNA, and amplified in a PCR. The PCR products are the template for further T7 in vitro transcription to produce rRNA fragments for the next round of selection. After a significant increase in binding, the fragments are cloned and sequenced. The SERF technique leads directly to the native binding site for the protein.