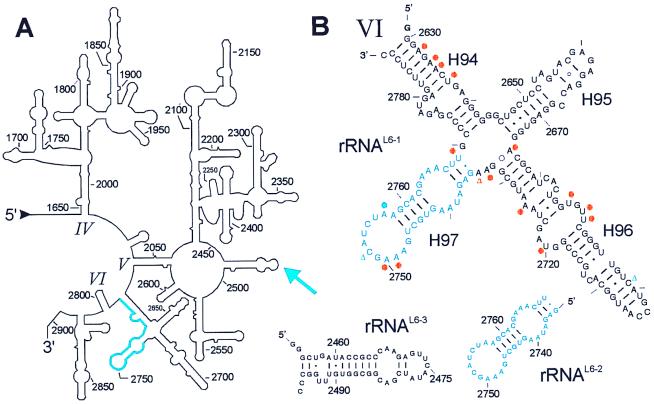
Figure 5.
Secondary structures of a part of the 23S rRNA and some fragments analyzed for binding to the ribosomal protein L6. (A) Secondary structure of the 3′ region of 23S rRNA with domain IV-VI (1647–2904; E. coli numbering; ref. 48). The rRNA fragment selected in the L6-SERF experiment is highlighted in blue. The blue arrow indicates a crosslink between L6 and helix H89 of domain V in 23S rRNA (31, 42). (B) rRNAL6-1 is a subdomain of domain VI with 161 nt comprising the sequence G-G2630-C2789 and including the functionally important sarcin/ricin stem loop (H95). The minimal L6-binding site is in blue. L6 induced changes of base reactivities against dimethylsulphate are indicated with a blue dot (protection) and blue triangles (enhancements) (41). Some bases are protected against base-specific chemical probing by protein L3 within rRNAL6-1 (orange dots, protection; triangle, enhancement; refs. 41 and 47). This fragment binds both L3 and L6. Other fragments used in this study are RNAL6-2 (G2735-U2769, 35 nt), the minimal fragment for L6 binding, and rRNAL6-3 (G-G2455-C2496-CC, 46 nt; helix H89) that originates from domain V of 23S rRNA and contains the crosslink site of L6. This fragment does not bind the protein.