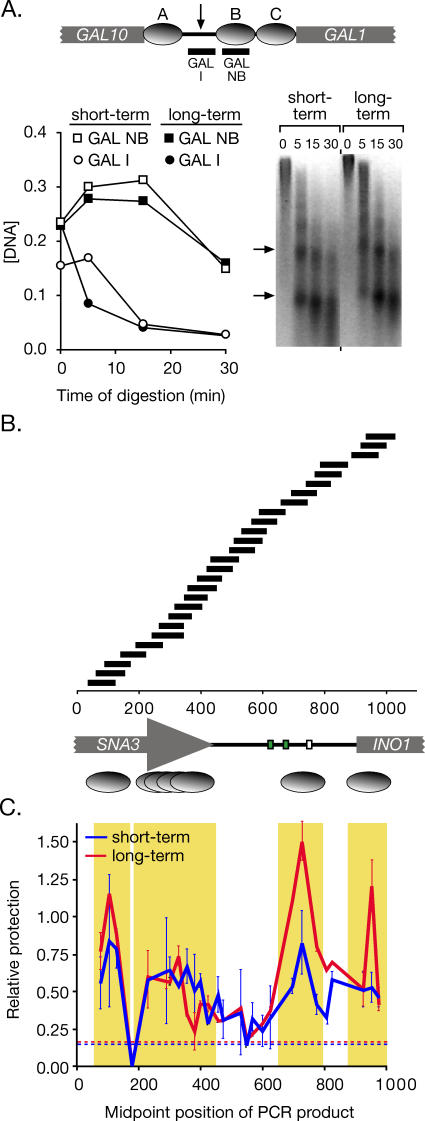
Figure 5. Nucleosome Positioning and Relative Occupancy in the Long-Term Repressed and Short-Term Repressed INO1 Promoters.
(A) Top panel: a map of three known, well-positioned nucleosomes, called “A,” “B,” and “C” (grey ovals) within the GAL1–10 promoter [33]. PCR products to monitor the concentration of sequences protected by nucleosome “B” (GAL NB) or sequences from the inter-nucleosomal region (arrow; GAL I) are indicated below. Bottom panel: DNA from either long-term repressed (open symbols) or short-term repressed (1 h; filled symbols) strain CRY1 was digested with micrococcal nuclease. The concentration of the templates for GAL NB (squares) and GAL I (circles) was determined relative to intact yeast genomic DNA by Q-PCR and plotted against different time points of digestion. Right: inverted image of ethidium bromide–stained gel of the digestion reactions.
(B) A map of the INO1 promoter and flanking sequences, along with the positions of PCR products quantified to analyze nuclease protection and nucleosome occupancy. Green boxes represent UASINO elements, and the white box represents the TATA box. Below the map, well-positioned nucleosomes are indicated as single ovals, and poorly positioned nucleosomes are indicated as an overlapping series.
(C) DNA from short-term and long-term repressed cells digested with micrococcal nuclease for 30 min (A) was analyzed by Q-PCR. Relative protection of the templates for each PCR product in (B) was calculated as a ratio of the concentration of the GAL NB template and mapped using the midpoint of the PCR product. Error bars represent standard error. The hatched lines represent the relative protection of the non-nucleosomal GAL I for each sample. Yellow boxes highlight protected sequences.