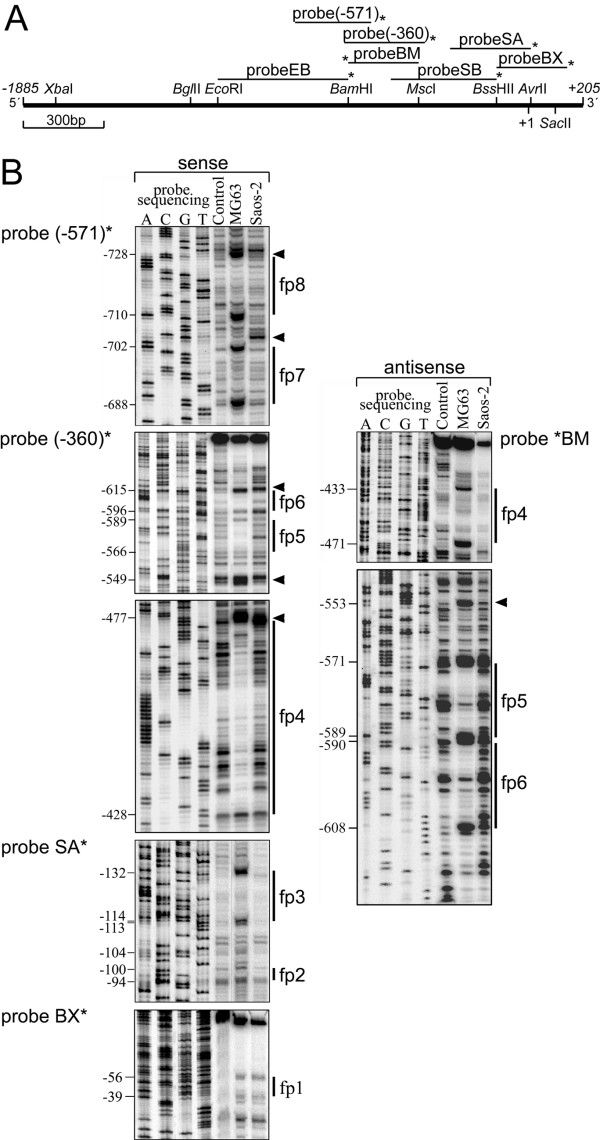
Figure 3.
In vitro DNaseI footprint analysis of protein-binding to the PDPN promoter. A) DNA probes used in DNaseI footprinting depicted above a restriction map of the -1885/+205 bp PDPN 5'-genomic region. These were: probeEB (bp -1169 to -646), probe(-571) (bp -857 to -571), probe(-360) (bp -666 to -360), probeBM (bp -649 to -385), probeSB (bp -498 to -85), probeSA (bp -261 to +31), and probeBX (bp -89 to +171). Asterisks indicate the positions of radioactive labeling as described in the experimental procedures section. Six of the probes analyze the coding DNA strand, whereas probe BM tracks the noncoding strand. B) DNaseI footprinting of probes using MG63 and Saos-2 nuclear cell extracts. End-labeled restriction fragments were incubated alone (Control) or with MG63 or Saos-2 cell nuclear extracts prior to digestion with DNase I. Footprinting was carried out in the sense and antisense direction. A sequencing reaction of the same region was run in parallel for exact size assignment of the protected regions. Protected sequences are indicated with single lines. Arrowheads denote sites hypersensitive to DNaseI.