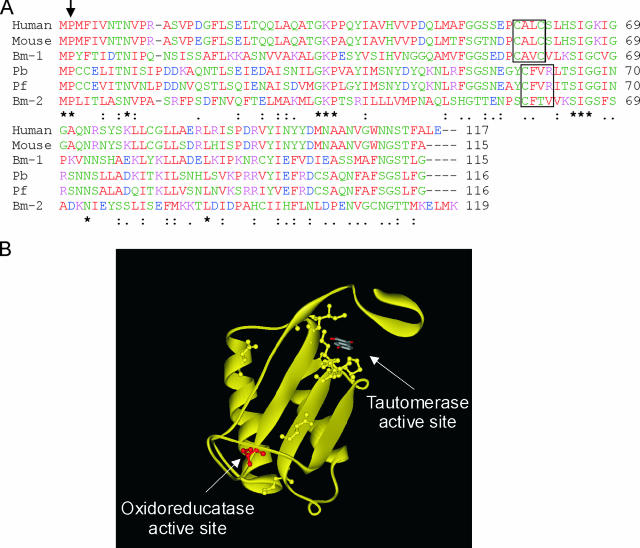
FIG. 1.
Sequence alignment of mammalian and parasite MIFs. (A) Sequence alignment of human, mouse, P. berghei (Pb), and P. falciparum (Pf) MIF homologues, as well as two Brugia malayi MIF homologues (Bm-1 and -2). The arrow indicates the N-terminal catalytic proline associated with tautomerase activity, and the area enclosed in a box is the CXXC motif associated with oxidoreductase activity in the mammalian MIFs and the parasite variants. Asterisks indicate identical residues; colons and dots indicate residues with high and low levels of similarity, respectively. (B) Ribbon representation of the huMIF crystal structure (RCSB PDB entry: 1CA7 [33]). For clarity, only one monomer of the trimer is shown. Arrows indicate the positions of the tautomerase and oxidoreductase active sites. The first cysteine of the CXXC oxidoreductase motif (C57 in huMIF) that is conserved in the parasite MIFs is red. Conserved residues in the various MIFs mainly cluster around the tautomerase active site and are shown in ball-and-stick form.