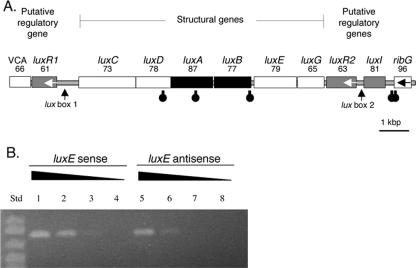
FIG. 2.
Bioinformatic and RT-PCR transcriptional analysis of the V. salmonicida lux region motifs. (A) The putative direction of transcription is from left to right unless indicated otherwise by an arrow. Black and gray highlighting denotes ORFs corresponding to luciferase genes and lux regulatory genes, respectively. The number listed below each gene name is the percentage of identity between the predicted amino acid sequence for the ORF and the sequence of the equivalent lux protein in V. fischeri strain MJ1. Stem-loop symbols below the ORFs represent the approximate locations of putative rho-independent transcriptional terminators. VCA is a homolog of the gene for the VCA0181 protein, a V. cholerae hypothetical protein with homologs found in both V. salmonicida and V. fischeri. (B) RT-PCR products of sense (lanes 1 to 4) and antisense (lanes 5 to 8) transcripts of luxE amplified from V. salmonicida total cellular RNA. The RNA extract was either undiluted (lanes 1 and 5), diluted 1:10 (lanes 2 and 6), diluted 1:100 (lanes 3 and 7), or diluted 1:1,000 (lanes 4 and 8). RT-PCR performed with no added RNA gave no product (data not shown). In lanes 1, 2, 5, and 6, a band matching the predicted 803-bp luxE product can be seen. Std, standard size markers; from top: 1,000, 900, 700, and 500 bp.