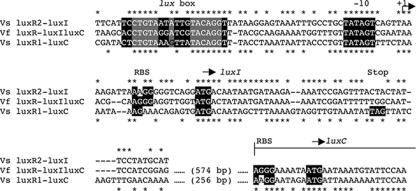
FIG. 4.
Sequence comparison of the intergenic regions between luxR2 and luxI of V. salmonicida (Vs luxR2-luxI), luxR and luxIC of V. fischeri MJ1 (Vf luxR-luxIluxC), and luxR1 and luxC of V. salmonicida (Vs luxR1-luxC). The V. fischeri luxR-luxIC and V. salmonicida luxR1-luxC sequences have been extended into their luxC ORFs. Identical nucleotides in the V. salmonicida luxR2-luxI and V. fischeri luxR-luxIC sequences are indicated with asterisks above the alignment. Identical nucleotides in the V. fischeri luxR-luxIC and V. salmonicida luxR1-luxC sequences are indicated with asterisks below the alignment. The highlighted sequences are based on motifs conserved around V. fischeri lux genes (17). These regions include a lux box, a −10 promoter region, and known or putative ribosome binding sites (RBS). Start (ATG) and stop (TAG) codons are also highlighted. Gray boxes highlight sequences required for a functional lux box in V. fischeri MJ1 (6).