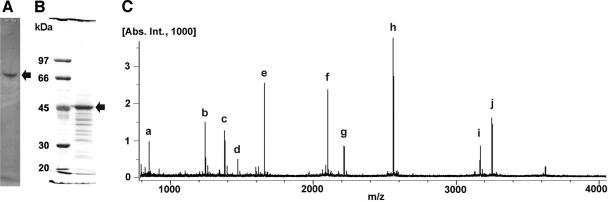
FIG. 1.
(A) Native PAGE of partially purified protein from Mesorhizobium sp. strain LUK showing the stained activity band (arrow). (B) SDS-PAGE of partially purified protein from native PAGE. The protein band of ca. 47 kDa (arrow) was analyzed further by mass spectrometry. (C) MALDI spectrum of the in-gel tryptic-digested 47-kDa protein from SDS-PAGE. One hundred images were collected to draw the cumulative chromatogram. Twenty peaks were selected from the identically observed peaks from the three independent experiments. The 10 highest peaks correspond to the signals for the peptides of residues 234 to 240 (a; AFLDLLR; M + H/z, 847.5036; calculated, 847.56), residues 396 to 404 (b; ELFFFHMLR; M + H/z, 1,239.6343; calculated, 1,239.57), residues 281 to 294 (c; YIGGGMSFGAFGGR; M + H/z, 1,376.6415; calculated, 1,376.53), residues 413 to 426 (d; GMYALSLEIADAGR; M + H/z, 1,466.7307; calculated, 1,466.61), residues 427 to 441 (e; DAFAEALADFIGEQR; M + H/z, 1,652.7914; calculated, 1,652.64), residues 55 to 73 (f; SILFHRPFPLVIAQGTGSR; M + H/z, 2,096.1763; calculated, 2,096.05), residues 141 to 162 (g; FTNSGTEANLMALATATAITGR; M + H/z, 2,211.1074; calculated, 2,211.00), residues 355 to 378 (h; IAVENQAPLQFTGLGSLGTIHFSR; M + H/z, 2,556.3568; calculated, 2,556.26), residues 306 to 335 (i; DGAFAHAGTFNNNILTMSAGHAALTQIYTR; M + H/z, 3,163.5377; calculated, 3,163.32), and residues 74 to 102 (j; FQDVDGHAYVNFLGEYTAGLFGHSHPVIR; M + H/z, 3,246.5755; calculated, 3,246.38). Peak b was analyzed by MALDI-MS/MS to get the internal peptide sequence.