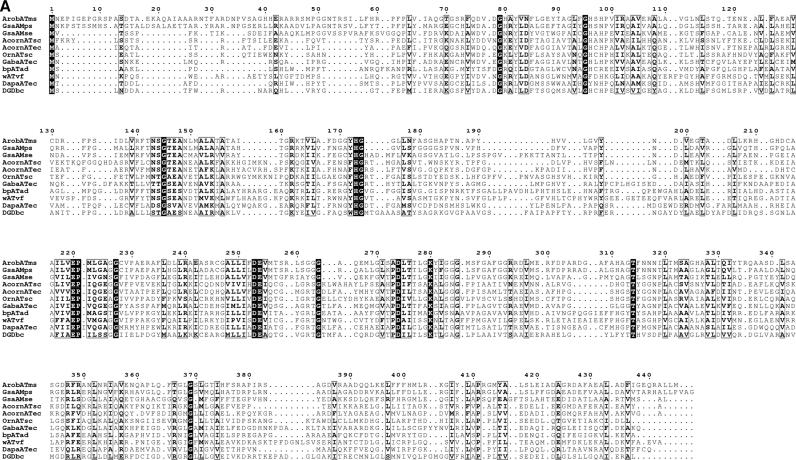
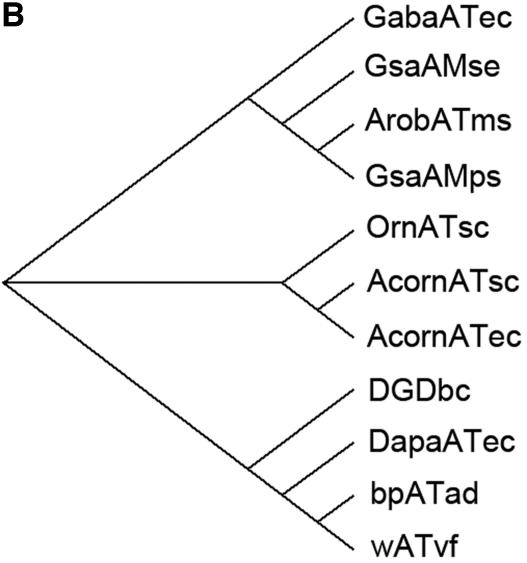
FIG. 3.
Sequence comparison with transaminase subgroup II enzymes. (A) Sequence alignment of the β-transaminase and other related proteins. Highly conserved residues are highlighted in black, and less strongly conserved residues are in gray boxes. Proteins: ArobATms, β-transaminase in this study; GsaAMps, glutamate-1-semialdehyde 2,1-aminomutase of Polaromonas sp. strain JS666 (gi, 91787361); GsaAMse, glutamate-1-semialdehyde 2,1-aminomutase of Synechococcus elongatus (gi, 581789); AcornATec, acetylornithine transaminase of Escherichia coli K-12 (gi, 16131238); AcornATsc, acetylornithine transaminase of Saccharomyces cerevisiae (gi, 6324432); OrnATsc, ornithine transaminase of Saccharomyces cerevisiae (gi, 6323470); GabaATec, 4-aminobutyrate transaminase of Escherichia coli K-12 (gi, 16130576); bpATad, β-alanine:pyruvate transaminase of Alcaligenes denitrificans (gi, 33086798); wATvf, ω-transaminase of Vibrio fluvialis (47); DapaATec, 7,8-diaminopelargonic acid synthase of Escherichia coli K-12 (gi, 16128742); DGDbc, 2,2-dialkylglycine decarboxylase of Burkholderia cepacia (gi, 729318). (B) A molecular phylogenetic tree. Abbreviations are as defined for panel A.