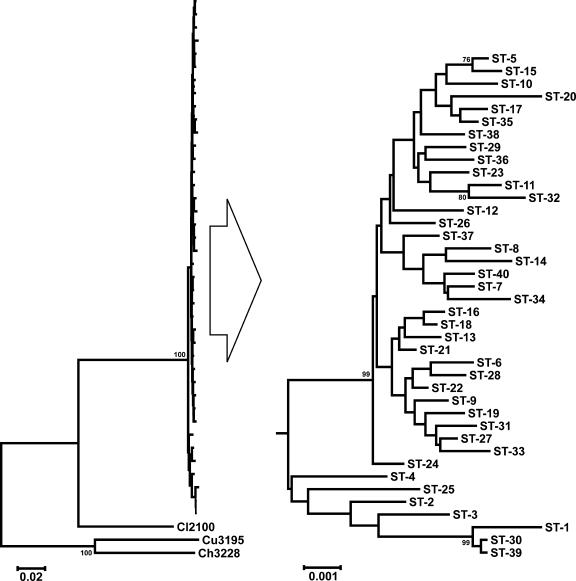
FIG. 2.
Dendrogram of Campylobacter insulaenigrae STs, including original isolates from the United Kingdom (ST-1 through ST-4). The dendrogram was constructed using the neighbor-joining algorithm and the Kimura two-parameter distance estimation method. Bootstrap values of >75%, generated from 500 replicates, are shown at the nodes. The scale bar represents substitutions per site. Three representative C. lari (Cl2100), C. upsaliensis (Cu3195), and C. helveticus (Ch3228) STs (using similar concatenated MLST alleles) are included for comparison. The C. insulaenigrae portion of the tree on the left is expanded on the right (large arrow) to improve clarity, since the scale of this portion of the tree with the related Campylobacter spp. included is too small to readily distinguish the C. insulaenigrae STs (note different scale bars).