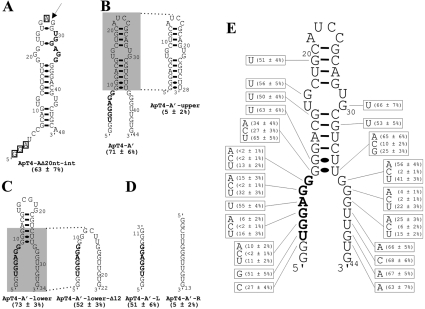
Figure 3. Determination of a minimal structure and important nucleotides for ApT4-A.
(A) Nucleotide sequence and proposed secondary structure of the deletion mutant ApT4-AΔ20nt-int. The binding activity is indicated in parentheses. The highly conserved sequence UGGAGG is shown in bold. The boxed nucleotides indicate bases that were deleted when designing ApT4-A' [see (B)]. The arrow indicates the position where the loop of ApT4-AΔ20nt-int was opened to produce ApT4-A'. (B) Nucleotide sequences and proposed secondary structures of ApT4-A' and ApT4-A'-upper. (C) Nucleotide sequences and proposed secondary structures of ApT4-A'-lower and ApT4-A'-lower-Δ12. (D) Nucleotide sequences and proposed secondary structures of ApT4-A'-L and ApT4-A'-R. (E) Nucleotide sequence and proposed secondary structure of ApT4-A'. All point mutations are indicated in the boxes along with the observed binding levels (averages from at least two independent experiments).