Abstract
We compared the results of two typing methods for 678 strains of methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. PCR-restriction fragment length polymorphism typing of the coagulase gene was a more reliable method than coagulase serotyping from the viewpoint of arbekacin resistance.
Molecular typing plays an important role in epidemiological studies of nosocomial infection, such as methicillin-resistant Staphylococcus aureus (MRSA) infection. Pulsed-field gel electrophoresis and multilocus sequence typing are considered the most discriminatory and reliable methods of typing, although they are technically complex, time consuming, and expensive. Coagulase serotyping is widely used in Japan in addition to conventional and genetic methods for distinguishing S. aureus strains. On the other hand, it has been reported that PCR-restriction fragment length polymorphism (RFLP) typing of the coagulase gene (coa) can be used to discriminate S. aureus strains on the basis of sequence variation within the 3′ end coding region of the gene (1, 2, 8). Arbekacin is effective, even in gentamicin-resistant MRSA strains, and has been used extensively in Japan since its approval as an anti-MRSA agent in 1990 (9, 10). We have studied arbekacin resistance in MRSA and recently reported the Japanese trend of arbekacin-resistant MRSA strains in the last two decades by using coa-RFLP typing (10). The results showed that arbekacin-resistant MRSA strains were distributed over only a few coa-RFLP types and remained at a low level. In this study, we assessed the clinical usefulness of coa-RFLP typing, especially from the viewpoint of arbekacin resistance, by comparing coagulase serotyping in both clinically isolated MRSA and clinically isolated methicillin-susceptible S. aureus (MSSA) in Japan.
Coagulase serotyping was performed with a coagulase-typing reagent kit (Denka Seiken Co. Ltd., Tokyo, Japan) according to the manufacturer's instructions. Based on the results, a total of 678 isolates, including 206 MSSA and 472 MRSA strains from unrelated clinical sources in Japan between 1979 and 2000 (4, 6, 7), were classified into nine serotypes, I to VIII and “other” (Table 1). MRSA strains were divided into serotypes I, II, III, IV, and VII, and most (82%) of the MRSA strains belonged to serotype II. A similar trend was previously observed in MRSA isolated in Japan (3). The serotype distribution of MSSA strains differed from that of MRSA strains. MSSA strains contained all serotypes. However, the majority of the MSSA strains were classified into serotypes II (28%) and VII (35%).
TABLE 1.
Correlation between the coagulase serotype and the coa-RFLP type in MRSA and MSSAa
| RFLP type | No. of strains with serotype
|
Total no. of strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | VIII | Other | ||
| XS15 | 2 (0/2) | 2 (0/2) | ||||||||
| SS01 | 1 (0/1) | 1 (0/1) | ||||||||
| SS21 | 1 (0/1) | 1 (1/0) | ||||||||
| S12 | 4 (1/3) | 2 (0/2) | 6 (1/5) | |||||||
| S15 | 1 (1/0) | 1 (1/0) | ||||||||
| S23 | 14 (6/8) | 14 (6/8) | ||||||||
| S24 | 2 (1/1) | 2 (1/1) | ||||||||
| S25 | 8 (0/8) | 8 (0/8) | ||||||||
| M11 | 13 (0/13) | 13 (0/13) | ||||||||
| M12 | 23 (1/22) | 10 (0/10) | 33 (1/32) | |||||||
| M22 | 29 (16/13) | 1 (1/0) | 30 (17/13) | |||||||
| M23 | 3 (3/0) | 3 (3/0) | ||||||||
| M31 | 12 (3/9) | 12 (3/9) | ||||||||
| M41 | 1 (0/1) | 1 (0/1) | ||||||||
| L01 | 1 (0/1) | 17 (2/15) | 2 (0/2) | 20 (2/18) | ||||||
| L11 | 2 (2/0) | 2 (2/0) | ||||||||
| L16 | 10 (8/2) | 2 (0/2) | 1 (0/1) | 13 (8/5) | ||||||
| L21 | 371 (356/15) | 1 (0/1) | 7 (7/0) | 379 (363/16) | ||||||
| L22 | 42 (37/5) | 1 (1/0) | 43 (38/5) | |||||||
| L23 | 26 (0/26) | 26 (0/26) | ||||||||
| L24 | 12 (0/12) | 4 (0/4) | 16 (0/16) | |||||||
| L31 | 27 (24/3) | 27 (24/3) | ||||||||
| L35 | 1 (0/1) | 1 (0/1) | ||||||||
| L51 | 1 (0/1) | 1 (0/1) | ||||||||
| LL01 | 1 (0/1) | 1 (0/1) | ||||||||
| LL21 | 1 (0/1) | 1 (0/1) | ||||||||
| LL27 | 4 (1/3) | 4 (1/3) | ||||||||
| LL31 | 1 (0/1) | 1 (0/1) | ||||||||
| XL21 | 13 (0/13) | 13 (0/13) | ||||||||
| XL22 | 1 (0/1) | 1 (0/1) | 2 (0/2) | |||||||
| XXL41 | 1 (0/1) | 1 (0/1) | ||||||||
| Total | 10 (8/2) | 436 (389/47) | 34 (11/23) | 50 (37/13) | 25 (0/25) | 3 (0/3) | 90 (18/72) | 13 (0/13) | 17 (9/8) | 678 (472/206) |
Results are shown as no. of strains (no. of MRSA strains/no. of MSSA strains).
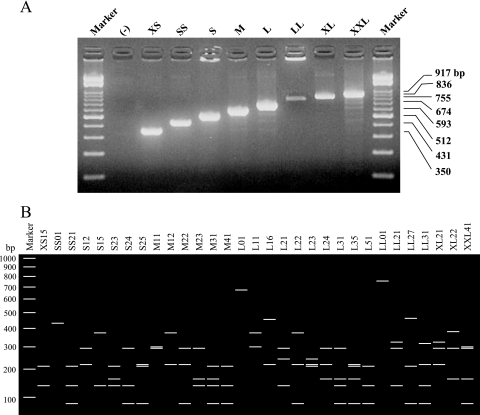
We next carried out coa-RFLP typing according to the method of Hookey et al., with some modifications (2). coa gene fragments were amplified from colonies grown on agar plates with the primers coaF (ATAGAGATGCTGGTACAGG) and coaR (GCTTCCGATTGTTCGATGC). As shown in Fig. 1A, PCR products of eight sizes were obtained from 678 isolates and named XS, SS, S, M, L, LL, XL, and XXL according to their sizes. The sizes of the products ranged from 350 to 917 bp in increments of 81 bp, reflecting the number of 81-bp repeat units contained in the coa gene. After digestion with AluI, 31 coa-RFLP types were detected and numbered to allow them to be distinguished from each other (Fig. 1B). The majority (77%) of MRSA strains belonged to type L21, indicating the spread of a specific type of MRSA in Japan. In contrast, there was no such tendency in MSSA. This result was in agreement with the results of serotyping.
FIG. 1.
Agarose gel electrophoresis pattern. (A) PCR amplification of coa genes. (B) Schematic representation of PCR-amplified coa genes digested with AluI.
A comparison of the results of the two typing methods may illuminate the advantages of coa-RFLP typing from the viewpoint of arbekacin resistance. Serotype II contains 11 coa-RFLP types, including type L31. Previously, we reported strain PRC104 as a highly arbekacin-resistant strain (128 μg/ml) (5). Although the typing data for this strain were not incorporated in Table 1 because of its history of isolation, the strain belongs to type L31 (unpublished data). This implies that this important coa-RFLP type cannot be recognized by coagulase serotyping alone. Types M22 and M31 are also important due to their high incidence of arbekacin-resistant strains (10). The coa-RFLP typing clearly distinguished type M22 or M31 isolates from other strains belonging to serotype VII or III, respectively. Thus, our results demonstrated that coa-RFLP typing has higher discriminatory power than coagulase serotyping and may be useful for discriminating groups that may be potential reservoirs of arbekacin-resistant MRSA.
Acknowledgments
We thank T. Okubo, S. Kondo, K. Kikuchi, Y. Arakawa, and A. Wada for MRSA strains and Y. Uehara for helpful discussion.
This work was supported by a grant from the Ministry of Health, Labor, and Welfare of Japan (Molecular Analysis of Drug-Resistant Bacteria and Establishment Rapid Identification Methods, no. H12-Shinkou-19).
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Goh, S.-H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 30:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ike, Y., Y. Arakawa, X. Ma, K. Tatewaki, M. Nagasawa, H. Tomita, K. Tanimoto, and S. Fujimoto. 2001. Nationwide survey shows that methicillin-resistant Staphylococcus aureus strains heterogeneously and intermediately resistant to vancomycin are not disseminated throughout Japanese hospitals. J. Clin. Microbiol. 39:4445-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishino, K., J. Ishikawa, Y. Ikeda, and K. Hotta. 2004. Characterization of a bifunctional aminoglycoside-modifying enzyme with novel substrate specificity and its gene from a clinical isolate of methicillin-resistant Staphylococcus aureus with high arbekacin resistance. J. Antibiot. 57:679-686. [DOI] [PubMed] [Google Scholar]
- 6.Kondo, S., Y. Ikeda, S. Hattori, M. Hamada, and T. Takeuchi. 1991. Susceptibility of methicillin-resistant Staphylococcus aureus to various antibiotics. Classification by aminoglycoside-modifying enzymes and antibiotics active against MRSA. Jpn. J. Antibiot. 44:1211-1215. [PubMed] [Google Scholar]
- 7.Piao, C., T. Karasawa, K. Totsuka, T. Uchiyama, and K. Kikuchi. 2005. Prospective surveillance of community-onset and healthcare-associated methicillin-resistant Staphylococcus aureus isolated from a university-affiliated hospital in Japan. Microbiol. Immunol. 49:959-970. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzkopf, A., and H. Karch. 1994. Genetic variation in Staphylococcus aureus coagulase genes: potential and limits for use as epidemiological marker. J. Clin. Microbiol. 32:2407-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabata, M., M. Shimizu, M. Araake, and H. Ogawa. 2003. Relationship between arbekacin-susceptibility and aminoglycoside-resistant gene of methicillin-resistant Staphylococcus aureus (MRSA). Jpn. J. Antibiot. 56:36-43. [PubMed] [Google Scholar]
- 10.Tsuchizaki, N., K. Ishino, F. Saito, J. Ishikawa, M. Nakajima, and K. Hotta. 2006. Trends of arbekacin-resistant MRSA strains in Japanese hospitals (1979 to 2000). J. Antibiot. 59:229-233. [DOI] [PubMed] [Google Scholar]