Abstract
To understand the role of mucosa-associated microbiota in the pathogenicity of ulcerative colitis (UC), paired biopsies were obtained during colonoscopy from the ulcerated and nonulcerated gut mucosa of 24 patients with UC. Denaturing gradient gel electrophoresis analysis was employed to profile the composition of the dominant bacteria (16S rRNA gene V3 region) and three important groups: lactobacilli, the Clostridium leptum subgroup, and Bacteroides spp. The Pearson coefficient was used to estimate similarities between the bacterial communities of the paired biopsies for each patient. The average similarity values of bacterial composition between the paired samples were 94.8 ± 3.8% for dominant bacteria, 59.9 ± 26.1% for lactobacilli, 79.2 ± 22.6% for the Clostridium leptum subgroup, and 88.7 ± 16.4% for Bacteroides spp. The data revealed that lactobacilli and the Clostridium leptum subgroup were significantly different between the ulcerated and the nonulcerated regions. It also was noted that for lactobacilli, the composition varied significantly between biopsy sites irrespective of the location of UC in the gut but that the composition of the Clostridium leptum subgroup showed significant differences between paired samples from UC in the rectum and not in the left colon. Localized dysbiosis of the mucosa-associated intestinal microflora, especially for lactobacilli and the Clostridium leptum subgroup, may be closely related to UC.
Ulcerative colitis (UC) is an acute and chronic inflammatory disease of the large bowel and is one of the two main forms of inflammatory bowel disease (IBD). The etiology of IBD is unknown, but animal models have shown that resident intestinal bacteria play an important role in the pathogenesis of this disease (27, 39).
Considerable effort has been expended on the study of intestinal bacteria in patients with inflammatory bowel disease. It has been reported that alterations of fecal microbiota present in IBD and the dominant bacteria are composed of unusual bacterial species compared with that of healthy subjects (36). Reduced diversity of the bacterial phylum Firmicutes in the fecal microbiota of Crohn's disease patients was observed (19) and, furthermore, a decreased number of lactobacilli, Clostridium leptum, and Clostridium coccoides in the fecal microflora, while an increased number of enterobacteria have been observed in patients with inflammatory bowel disease (3, 32, 36).
Since mucosa-associated bacteria induce a local immune response (24) and the mucosa-associated bacterial composition is significantly different in the feces (43), several more-recent studies have focused on the mucosa-associated bacteria in IBD patients. Overall, there is a larger population of aerobic and facultative-anaerobic bacteria in IBD patients but a decreased number of normal anaerobic bacteria (5, 18, 22).
Since there typically is a large person-to-person variation in the mucosa-associated bacteria populating the gut (43), it may be more relevant to compare the intestinal bacteria within the same individual (15). However, no significant differences between the amounts of bacteria from inflamed and noninflamed biopsies were observed in patients with IBD (38); in addition, the dominant microbiota did not differ qualitatively in Crohn's disease patients (31).
The differences among the intestinal bacterial community adherent to the ulcerated and nonulcerated mucosa in the same patient with ulcerative colitis have not been explored in detail. In this work, we employed denaturing gradient gel electrophoresis (DGGE) to profile the biodiversity of the dominant bacteria and three important bacterial groups in paired biopsies from patients with UC to search for patterns associated with the progression of this important disease.
MATERIALS AND METHODS
Volunteers and colonoscopy biopsies.
Twenty-four ulcerative colitis patients were involved in this study (Table 1). One patient had cecal UC, 9 had left-colonic UC, 1 had rectosigmoid UC, and 13 had rectal UC. Diagnosis of UC was confirmed by histology at the time of colonoscopy, and all patients had active mild to moderate ulcerative colitis (26). Nine of the patients received therapy with mesalazine (5-aminosalicylic acid), three of them received corticosteroids, and none of them had received antibiotics within the previous 4 weeks. Colonic cleansing was performed with polyethylene glycol-electrolyte powder (WanHe Pharmaceutical Co., Ltd., Shenzhen, China). Two biopsies were taken from each patient after the colonic evacuation. One biopsy was collected from a macroscopic ulcer or an erosion-mucosal site, which is referred to as an ulcerated mucosa in this study, and the other was from adjacent (about 5 cm away) mucosa that appeared normal macroscopically, which is referred to as nonulcerated mucosa. In order to minimize contamination, different colonoscopy jaws were used to obtain the ulcerated and the nonulcerated mucosa biopsies within a given individual. The volunteers underwent routine diagnostic colonoscopy, and biopsies were included in this procedure, so this study did not add extra risk to the procedure. Informed consent was obtained from each patient.
TABLE 1.
Information about the patients
| Sampling position | No. of patients | Sex (male/female) | Mean age (yr) (range) | Treatment
|
|
|---|---|---|---|---|---|
| Corticosteroid | Mesalazine | ||||
| Cecum | 1 | 1/0 | 16 (16) | 0 | 1 |
| Left colon | 9 | 4/5 | 47 (34-72) | 1 | 3 |
| Rectosigmoid | 1 | 0/1 | 38 (38) | 0 | 0 |
| Rectum | 13 | 4/9 | 46 (19-70) | 2 | 5 |
| Total | 24 | 9/15 | 40 (16-72) | 3 | 9 |
DNA extraction and PCR amplification.
Biopsy samples (0.5 to 1 mg) were suspended in 450 μl 0.05 M potassium phosphate buffer (pH 7.0) in a 2-ml Eppendorf tube. Samples were vortexed for 2 min at maximum speed using a Mo Bio vortex (model G-560E). This suspension was incubated at 55°C for 1 h with 10 μl proteinase K solution (20 mg/ml) and 50 μl 10% sodium dodecyl sulfate, followed by tissue dissociation in 150 μl phenol (pH 7.5) using a minibead beater (Biospec Products, Bartlesville, OK). This step was conducted three times and each time at the maximum speed for 1 min, followed by placement on ice for 1 min.
After dissociation, 150 μl chloroform-isoamyl (vol/vol, 24:1) was added and centrifuged at 15,000 × g for 10 min. The supernatant was subjected to extraction with an equal volume of phenol, followed by phenol-chloroform-isoamyl (chloroform vol/isoamyl vol, 24:1; phenol vol/chloroform-isoamyl vol, 1:1) and then chloroform-isoamyl (vol/vol, 24:1). DNA was precipitated with two volumes of ethanol and 1/10 volume of sodium acetate (3 M, pH 5.2) and collected by centrifugation (15,000 × g for 10 min), air dried, and dissolved in 100 μl sterile TE (Tris-EDTA) buffer. RNA was digested by adding 3 μl RNase (20 mg/ml) at 37°C for 20 min.
Since the biopsy sample was too small and the DNA extracted from the biopsy contained a higher amount of eukaryotic DNA than bacterial DNA, nested PCR was employed in this study to ensure specificity in the amplification of 16S rRNA genes. Primers 27F and 1492R (7, 14), which were designed based on conserved bacterial regions at the 5′ and 3′ ends of the 16S rRNA gene, were used to amplify the near-full-length gene. The 25-μl reaction mixture contained 2.5 μl of 10× PCR buffer (Mg2+ free), 2 μl of a 25 mM concentration of each deoxynucleoside triphosphate (dNTP) mixture, 0.625 U Ex Taq DNA polymerase (Takara, Dalian, China), and 25 pmol of each primer. Amplification was performed by initial denaturation at 94°C for 4 min, followed by 20 cycles consisting of 94°C for 45 s, 55°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 6 min. PCR amplification was performed with a thermocycler PCR system (PCR Sprint; Thermo Electron Corp., United Kingdom). The amplification products of the 16S rRNA gene were used as the templates in the next amplification.
Dominant bacteria were characterized by amplifying the V3 region of the 16S rRNA gene, and group-specific DGGE analysis was used for lactobacilli, the Clostridium leptum subgroup, and the Bacteroides spp. For the 16S rRNA gene V3 region amplification, the 25-μl reaction mixture contained 2.5 μl of 10× PCR buffer (Mg2+ free), 2 μl of a 25 mM concentration of each dNTP, 0.625 U of Ex Taq DNA polymerase, and 12.5 pmol of each primer (20). The samples were amplified in a thermocycler PCR system by a touchdown PCR protocol (20).
Lactobacilli were detected by the specific primers Lac 1 and Lac 2GC that target the 16S rRNA gene (13, 40) to amplify a 384-bp product; the Clostridium leptum subgroup was amplified with group-specific primers Clept-F and Clept-GC-R3, producing a 279-bp product (34). Bacteroides genus-specific primers Bfr-F and Bfr-G-CR (23), which target the 16S rRNA gene, were used to amplify a 270-bp product. The PCR mixture (25 μl) contained 0.625 U of Ex Taq DNA polymerase, 10× PCR buffer (Mg2+ free), 20 pmol of each primer, and a 50 mM concentration of each dNTP. Amplification was carried out in a thermocycler PCR system. The amplifications were performed as previously described (23, 40). Five microliters of each PCR product was checked by electrophoresis on 1% (wt/vol) agarose gels.
DGGE profiling and data analysis.
Parallel DGGE was performed with a Dcode System apparatus (Bio-Rad) as described by the manufacturer. Amplification products were separated on 8% (wt/vol) polyacrylamide gels. Electrophoresis was performed in 1× Tris-acetate-EDTA (TAE) buffer at a constant voltage of 200 V and a temperature of 60°C for 200 min. The DNA bands were stained with SYBR green I (Amresco) and photographed using a UVI gel documentation system (UVItec, Cambridge, United Kingdom).
For 16S rRNA gene V3 region analysis, the DGGE gel contained a linear 35% to 55% denaturing gradient (100% denaturant corresponds to 7 M urea and 40% deionized formamide). For the lactobacilli and the Clostridium leptum subgroup analysis, the DGGE gel contained a linear 28% to 48% denaturing gradient, and for Bacteroides genus-specific analysis, the DGGE gel contained a linear 22.5% to 45% denaturing gradient.
Gelcompare II software (Applied Maths, Kortrijk, Belgium) was used for gel analysis, and the similarity score was calculated using the Pearson correlation. The methodological bias caused by the DNA extraction, PCR amplification, and DGGE analysis was estimated by performing these whole procedures on four samples twice (15), and the average similarity value of these four pairs of replicated samples was 93.8 ± 4.2%. In this study, the DGGE patterns with similarity values higher than 93.8% were not significantly different.
Bacterial diversity in the ulcerated site and the nonulcerated site was evaluated as the band numbers of DGGE profiles (32) and weighted diversity according to Shannon and Weaver (22, 33).
All data sets were expressed as the means ± standard deviations. Student's t test was applied for the comparison of variables with normal distribution, and the Wilcoxon signed-rank test was used for nonnormal distributions.
RESULTS
Biopsy samples and PCR amplification.
Forty-eight bacterial DNA samples were extracted from the ulcerated and nonulcerated mucosa of 24 patients diagnosed with ulcerative colitis. Since amplification of some DNA samples did not yield a positive result, 19 subjects were involved in lactobacilli and Clostridium leptum subgroup analyses. Seventeen subjects were involved in the Bacteroides genus-specific DGGE analysis.
Comparisons of bacterial community structure within an individual.
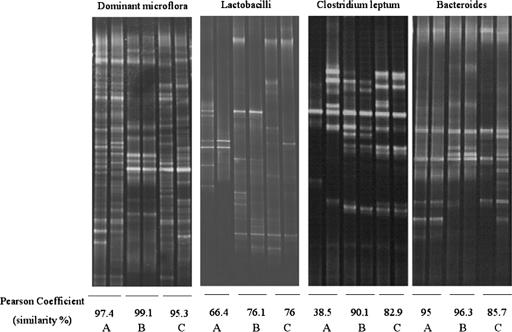
The DGGE patterns from three representative individuals are shown in Fig. 1. The DGGE patterns indicated that the bacterial composition varied from person to person. When the microbiota biodiversity within a given individual was considered, the dominant bacterial composition within each patient showed a high similarity. The similarity value ranged from 79.9% to 99.1%. The mean similarity percentage was 94.8 ± 3.8%, and this value was higher than 93.8%, which indicated that the dominant adherent bacterial composition was similar between the two mucosal sites.
FIG. 1.
DGGE patterns of the mucosa-associated microbiota in three individual patients, A, B, and C. A Pearson coefficient (percentage of similarity) was calculated between the two biopsy sites from each individual. The left lane for each individual reflected the bacteria from the ulcerated mucosa and the right lane reflected the nonulcerated mucosa.
For lactobacillus analysis, the similarity value ranged from 13.6% to 96.6% and the mean value was 59.9 ± 26.1%. The mean similarity value for the Clostridium leptum subgroup analysis was 79.2 ± 22.6% and the similarity percentage ranged from 27.6% to 98.8%. The similarity value for Bacteroides genus-specific DGGE patterns ranged from 37.2% to 99.3%, and the mean value was 88.7 ± 16.4%.
When the results of all patients were taken into account, the similarity values for lactobacilli and the Clostridium leptum subgroup showed significant reduction compared with that of the dominant bacteria (P < 0.01), while the similarity values for Bacteroides did not show a significant difference (Table 2). These results indicated that the dominant bacteria were more similar between two adjacent biopsies within the same intestinal region, while for other groups, especially for lactobacilli and the Clostridium leptum group, the bacterial composition varied significantly.
TABLE 2.
Average similarity coefficients between the mucosa-associated bacteria in ulcerated and nonulcerated positions from 24 ulcerative colitis patientsa
| Position | Coefficient (%) of similarity forb:
|
|||
|---|---|---|---|---|
| Dominant bacteria (n = 24) | Lactobacilli (n = 19) | Clostridium leptum (n = 19) | Bacteroides spp. (n = 17) | |
| Cecum | 96.4 (n = 1) | 18.3 (n = 1) | 92.1 (n = 1) | 87.3 (n = 1) |
| Left colon | 95.8 ± 0.8 (n = 9) | 60.4 ± 26.1 (n = 7)* | 81.5 ± 20.2 (n = 8) | 88.2 ± 15.4 (n = 6) |
| Rectosigmoid | 79.9 (n = 1) | 55.7 (n = 1) | 93.0 (n = 1) | 94.1 (n = 1) |
| Rectum | 95.2 ± 2.7 (n = 13) | 64.1 ± 26.3 (n = 10)** | 74.2 ± 26.8 (n = 9)* | 88.5 ± 19.6 (n = 9) |
| Total | 94.8 ± 3.8 (n = 24) | 59.9 ± 26.1 (n = 19)** | 79.2 ± 22.6 (n = 19)** | 88.7 ± 16.4 (n = 17) |
Values shown are percentages ± standard deviations.
*, P < 0.05 compared with the dominant bacteria. **, P < 0.01 compared with the dominant bacteria.
The bacterial diversity in ulcerated and nonulcerated sites was evaluated as band numbers and weighted diversity. No significant differences in bacterial diversity between the two mucosal sites were observed when we focused on the dominant bacteria and these three groups (data not shown).
Comparisons of bacterial composition from separate compartments of the lower intestine.
Four intestinal compartments, the cecum, the left colon, the rectosigmoid, and the rectum, were involved in this study, and most patients had ulcerative colitis localized to the left colon and rectum. The similarity percentages for the dominant bacteria and three groups were compared among the patients with left-colonic UC and rectal UC. The results indicated that in left-colonic UC, the average similarity index of only lactobacilli between the two biopsies (60.4 ± 26.1%) was decreased significantly more than that of the dominant bacteria (95.8 ± 0.8%; P < 0.05). In rectal UC, the average similarity index of both lactobacilli (64.1 ± 26.3%) and the Clostridium leptum subgroup (74.2 ± 26.8%) was reduced significantly compared with that of dominant bacteria (95.2 ± 2.7%, P < 0.01) (Table 2). These results suggested that different diseased regions might have differences in the type and extent of the bacterial community alteration.
DISCUSSION
In this work, DGGE patterns with a similarity value higher than 93.8% were thought of as having no differences, and a similar percentage also was regarded as the positive cutoff of the similarity value from previous reports (15, 31). In this study, the average similarity value of the dominant bacteria between two adjacent biopsies was higher than the positive cutoff of our study, and this meant that the dominant bacterial composition at the two biopsy sites was similar. Due to the host specificity of the intestinal bacteria and the limited sample size and bias produced during the sample collection and processing, the positive cutoff for the three groups was not titrated with replicate samples; instead, the similarities for the three groups between the paired samples in each patient were statistically analyzed against the similarity value of the dominant bacteria within the same host.
In order to avoid amplification of eukaryotic DNA from the biopsy samples, nested PCR was performed. In the first step, a nearly complete 16S rRNA gene fragment was amplified using the universal primer pair 27F/1492R. The product obtained was used as a template for a second amplification with dominant bacteria and three group-specific primers. Nested PCR can improve the specificity and make it possible to characterize the diversity of bacteria in low numbers from mixed microbial communities.
The mucosa-associated bacterial composition is host specific and significantly different from that of fecal bacteria (43), and the composition also is stable over a period of time (22). This suggested that the mucosa-associated bacterial composition alteration found in this study was not due to transient factors.
Dominant bacteria were found to be relatively stable when nonulcerated tissues were collected along the distal intestine (15), and the dominant bacteria from ulcerated and nonulcerated biopsies in Crohn's disease are similar (31). In this study, the universal primer-based DGGE analysis indicated that the dominant bacterial structures of ulcerated and the nonulcerated tissue showed high similarity. Due to the complexity of intestinal bacteria, the resolution of the universal primer-based analysis was not enough to reflect the bacterial composition in detail.
More recently, group-specific PCR-DGGE has been developed for important groups such as Bacteroides and the Clostridium leptum subgroup to increase the resolution and sensitivity of this technology for evaluating gut ecology (23, 34). This method allows the visualization of the bacterial group of interest against a highly diverse background (2, 35). In our study, group-specific DGGE analysis indicated that the bacterial composition in ulcerated and nonulcerated biopsies within the same intestinal segment showed lower similarities than those of the dominant bacteria. Furthermore, great intraindividual variations existed. Among the bacteria we investigated, the composition of lactobacilli and the Clostridium leptum subgroup varied significantly between the ulcerated and the nonulcerated areas within the same individual.
Lactobacilli are thought of as beneficial to the host, and many studies have demonstrated that some strains of lactobacilli can reduce the severity and maintain the remission of this disease (1). A significant decrease in the number of lactobacilli is found in the mucosa of patients with inflammatory bowel disease (9, 22), and it has been suggested that the changing condition in the intestine may influence the lactobacillus composition (43). We found that the composition of lactobacilli adhered to the ulcerated and nonulcerated tissue from within the same patient varied greatly and may be caused by differences in the physiological status of the mucosa.
The C. leptum subgroup (cluster IV) (4) is one of the most predominant populations of human fecal microflora (12, 37), which contains a large number of butyrate-producing bacteria (25, 28). The bacteria's metabolic activities have a significant effect on the health of the human colon. It has been reported that a decreased fecal population level of the Clostridium leptum subgroup is observed in patients with inflammatory bowel disease (36). When we compared the bacteria adhered to the ulcerated mucosa and the nonulcerated mucosa, we found that the composition of the Clostridium leptum subgroup differed significantly in rectal UC but not in left-colonic UC. It has been demonstrated that protein can be degraded into branched chain fatty acids by many Clostridia spp. (8) and that the breakdown of proteins by the microbiota becomes more important in the distal bowel than in the proximal region (17). Therefore, the results from those studies suggest that change in the mucosal physiology in the rectum may have a greater influence on the composition of the Clostridium leptum subgroup than in the left colon.
Bacteroides is the most dominant part of the normal indigenous flora in the human gut. It makes up more than 25% of bacteria in human fecal flora (10, 37, 41, 42). These bacteria are significant contributors to the metabolism, nutrition, and health of humans and animals; some Bacteroides species frequently are found in clinical infections and are thought to be opportunistic microorganisms (6, 21, 30). The role of Bacteroides spp. in inflammatory bowel disease has been explored in many investigations. Several studies reported an increase of Bacteroides spp. population level in IBD by the use of both culture-dependent and culture-independent methods (11, 16, 29, 38). However, Conte et al. found a decreased population level of anaerobic organisms, especially Bacteroides vulgatus (5). The age difference among the individuals involved in these studies may cause these discrepant results. Moreover, these studies focused on the population level of Bacteroides spp., while the present study assessed the biodiversity of this genus. Our results indicated that Bacteroides spp. composition did not differ markedly between the two biopsy sites within the same individual.
In summary, the composition of lactobacilli and that of Clostridium leptum were altered between the ulcerated and the nonulcerated biopsy sites, and the bacterial structure alteration was host specific. These results suggest that these bacteria may be closely related to UC and that this alteration may be caused by the differential physiology of the intestinal mucosa. The potential roles of lactobacilli and Clostridium leptum in the etiology of ulcerative colitis should be more closely evaluated in future research.
Acknowledgments
We thank all the volunteers involved in this study.
This work was supported by a grant from the National Natural Science Foundation of China (30370031).
Footnotes
Published ahead of print on 6 December 2006.
This work was performed at the Laboratory of Molecular Microbial Ecology and Ecogenomics, Shanghai Center for Systems Biomedicine, College of Life Science and Biotechnology, Shanghai Jiao tong University, 200240 Shanghai, China.
REFERENCES
- 1.Bibiloni, R., R. N. Fedorak, G. W. Tannock, K. L. Madsen, P. Gionchetti, M. Campieri, C. De Simone, and R. B. Sartor. 2005. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 100:1539-1546. [DOI] [PubMed] [Google Scholar]
- 2.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullock, N. R., J. C. Booth, and G. R. Gibson. 2004. Comparative composition of bacteria in the human intestinal microflora during remission and active ulcerative colitis. Curr. Issues Intest. Microbiol. 5:59-64. [PubMed] [Google Scholar]
- 4.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 5.Conte, M. P., S. Schippa, I. Zamboni, M. Penta, F. Chiarini, L. Seganti, J. Osborn, P. Falconieri, O. Borrelli, and S. Cucchiara. 2006. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 55:1760-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerden, B. I. 1994. Virulence factors in anaerobes. Clin. Infect. Dis. 184:S253-S259. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsden, S. R., and M. G. Hilton. 1978. Volatile acid production from threonine, valine, leucine and isoleucine by clostridia. Arch. Microbiol. 117:165-172. [DOI] [PubMed] [Google Scholar]
- 9.Fabia, R., A. Ar'Rajab, M. L. Johansson, R. Andersson, R. Willen, B. Jeppsson, G. Molin, and S. Bengmark. 1993. Impairment of bacterial flora in human ulcerative colitis and experimental colitis in the rat. Digestion 54:248-255. [DOI] [PubMed] [Google Scholar]
- 10.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giaffer, M. H., C. D. Holdsworth, and B. I. Duerden. 1991. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J. Med. Microbiol. 35:238-243. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi, H., M. Sakamoto, and Y. Benno. 2002. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46:535-548. [DOI] [PubMed] [Google Scholar]
- 13.Kuhbacher, T., S. J. Ott, U. Helwig, T. Mimura, F. Rizzello, B. Kleessen, P. Gionchetti, M. Blaut, M. Campieri, U. R. Folsch, M. A. Kamm, and S. Schreiber. 2006. Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis. Gut 55:833-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY.
- 15.Lepage, P., P. Seksik, M. Sutren, M. F. de la Cochetiere, R. Jian, P. Marteau, and J. Dore. 2005. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11:473-480. [DOI] [PubMed] [Google Scholar]
- 16.Lucke, K., S. Miehlke, E. Jacobs, and M. Schuppler. 2006. Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J. Med. Microbiol. 55:617-624. [DOI] [PubMed] [Google Scholar]
- 17.Macfarlane, G. T., G. R. Gibson, E. R. Beatty, and J. H. Cummings. 1992. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol. Ecol. 101:81-88. [Google Scholar]
- 18.Macfarlane, S., E. Furrie, J. H. Cummings, and G. T. Macfarlane. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38:1690-1699. [DOI] [PubMed] [Google Scholar]
- 19.Manichanh, C., L. Rigottier-Gois, E. Bonnaud, K. Gloux, E. Pelletier, L. Frangeul, R. Nalin, C. Jarrin, P. Chardon, P. Marteau, J. Roca, and J. Dore. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers, L. L., D. S. Shoop, L. L. Stackhouse, F. S. Newman, R. J. Flaherty, G. W. Letson, and R. B. Sack. 1987. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J. Clin. Microbiol. 25:2330-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott, S. J., M. Musfeldt, D. F. Wenderoth, J. Hampe, O. Brant, U. R. Folsch, K. N. Timmis, and S. Schreiber. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang, X., D. Ding, G. Wei, M. Zhang, L. Wang, and L. Zhao. 2005. Molecular profiling of Bacteroides spp. in human feces by PCR-temperature gradient gel electrophoresis. J. Microbiol. Methods 61:413-417. [DOI] [PubMed] [Google Scholar]
- 24.Poxton, I. R., R. Brown, A. Sawyerr, and A. Ferguson. 1997. Mucosa-associated bacterial flora of the human colon. J. Med. Microbiol. 46:85-91. [DOI] [PubMed] [Google Scholar]
- 25.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 26.Rachmilewitz, D. 1989. Coated mesalazine (5-aminosalicylic acid) versus sulphasalazine in the treatment of active ulcerative colitis: a randomised trial. Br. Med. J. 298:82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rath, H. C., H. H. Herfarth, J. S. Ikeda, W. B. Grenther, T. E. Hamm, Jr., E. Balish, J. D. Taurog, R. E. Hammer, K. H. Wilson, and R. B. Sartor. 1996. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J. Clin. Investig. 98:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert, C., and A. Bernalier-Donadille. 2003. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol. Ecol. 46:81-89. [DOI] [PubMed] [Google Scholar]
- 29.Ruseler-van Embden, J. G., and H. C. Both-Patoir. 1983. Anaerobic Gram-negative faecal flora in patients with Crohn's disease and healthy subjects. Antonie Leeuwenhoek 49:125-132. [DOI] [PubMed] [Google Scholar]
- 30.Sack, R. B., M. J. Albert, K. Alam, P. K. Neogi, and M. S. Akbar. 1994. Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. J. Clin. Microbiol. 32:960-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seksik, P., P. Lepage, M. F. de la Cochetiere, A. Bourreille, M. Sutren, J. P. Galmiche, J. Dore, and P. Marteau. 2005. Search for localized dysbiosis in Crohn's disease ulcerations by temporal temperature gradient gel electrophoresis of 16S rRNA. J. Clin. Microbiol. 43:4654-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seksik, P., L. Rigottier-Gois, G. Gramet, M. Sutren, P. Pochart, P. Marteau, R. Jian, and J. Dore. 2003. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shanon, C. W., and W. Weaver. 1963. Mathematical theory of communication. University of Illinois Press, Urbana.
- 34.Shen, J., B. Zhang, G. Wei, X. Pang, H. Wei, M. Li, Y. Zhang, W. Jia, and L. Zhao. 2006. Molecular profiling of the Clostridium leptum subgroup in human fecal microflora by PCR-denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 72:5232-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol, H., P. Seksik, L. Rigottier-Gois, C. Lay, P. Lepage, I. Podglajen, P. Marteau, and J. Dore. 2006. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 12:106-111. [DOI] [PubMed] [Google Scholar]
- 37.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swidsinski, A., A. Ladhoff, A. Pernthaler, S. Swidsinski, V. Loening-Baucke, M. Ortner, J. Weber, U. Hoffmann, S. Schreiber, M. Dietel, and H. Lochs. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44-54. [DOI] [PubMed] [Google Scholar]
- 39.Taurog, J. D., J. A. Richardson, J. T. Croft, W. A. Simmons, M. Zhou, J. L. Fernandez-Sueiro, E. Balish, and R. E. Hammer. 1994. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 180:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, K. H., J. S. Ikeda, and R. B. Blitchington. 1997. Phylogenetic placement of community members of human colonic biota. Clin. Infect. Dis. 25:S114-S116. [DOI] [PubMed] [Google Scholar]
- 43.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. L. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]