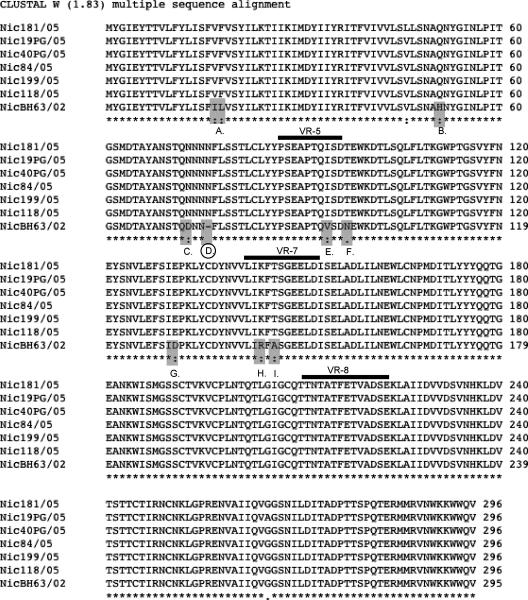
FIG. 4.
Amino acid alignment using ClustalW, version 1.8, with default parameters on the European Bioinformatics Institute server of the rotavirus VP7 gene showing the number of amino acid substitutions in the 2005 Nicaraguan isolates compared to the isolate from 2002. The substitutions occurring at positions A to I are all conserved or semiconserved. The three important antigenic sites (VR-5, aa 86 to 101; VR-7, aa 142 to 152; and VR-8, aa 211 to 223) are underlined. The location of the asparagine insert at position 76 is indicated with a circle. *, conserved residue; :, position with conserved substitutions; ., position with semiconserved substitutions.