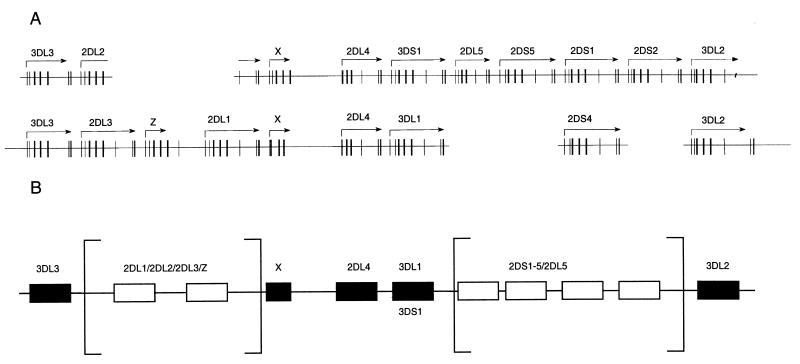
Figure 3.
(A) Differences in the organization of the KIR region in two different haplotypes. On the top line is the plot of the KIR region from 1060P112 (Fig. 2A). Partial sequence was obtained from PACs 72N61, 78E41, 1015E91, and 1015 M91 for the other haplotype (Fig. 1) by using PCR as well as comparison with other data (see Fig. 1). The gene distances are not to scale and have been exaggerated to display contiguity, suggested by sequence comparisons. Thus, 2DL2 is shown as a composite of two genes on the second haplotype, namely 2DL3 and 2DL1. Overall, the genes are so similar that the precise lineup of the alleles remains uncertain, but 3DS1 and 3DL1 are shown as alleles, as supported by other data (see below and ref. 16). (B) Framework genes and variable bubbles in the KIR region. Comparison of the gene organization data in Fig. 2A, from the two haplotypes, with other data (16) is consistent with invariant framework loci, flanking regions where the gene number shows marked flexibility, indicated as open boxes flanked by square boxes. A similar model has been proposed to explain (mostly interspecies) gene expansion/contraction in the MHC, proposing independent expansion of class I genes within a framework of ancestral loci (48).