Abstract
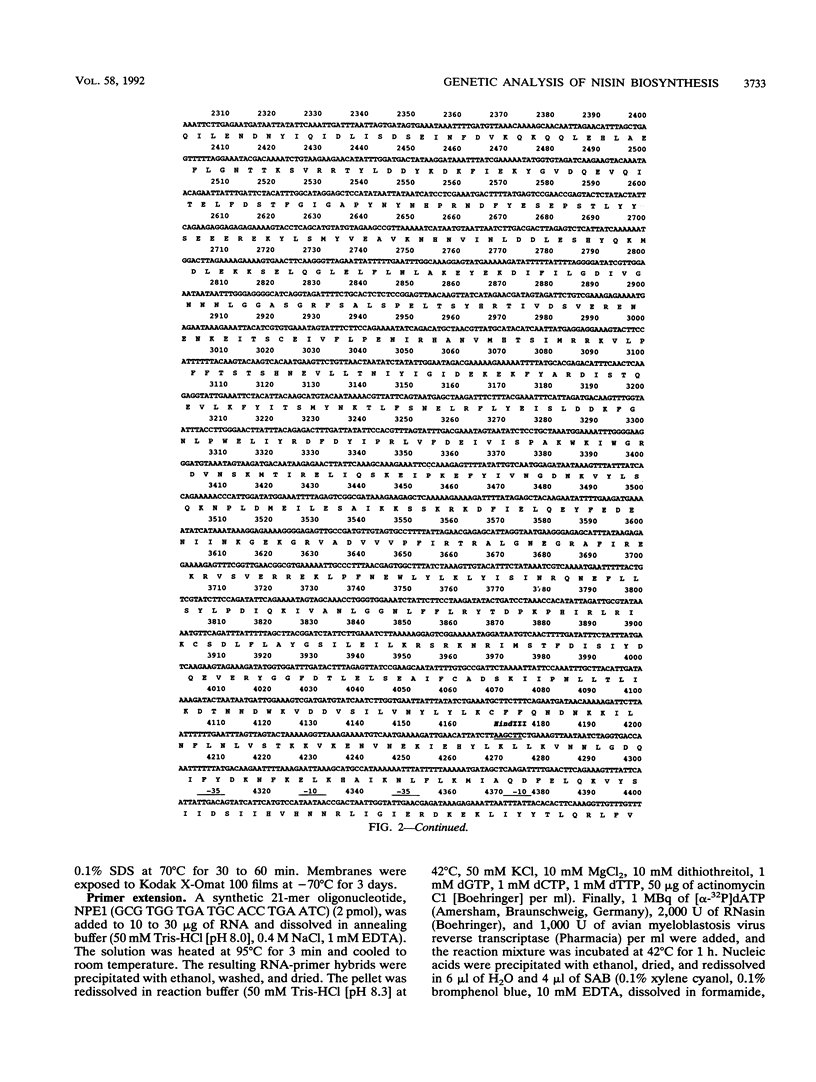
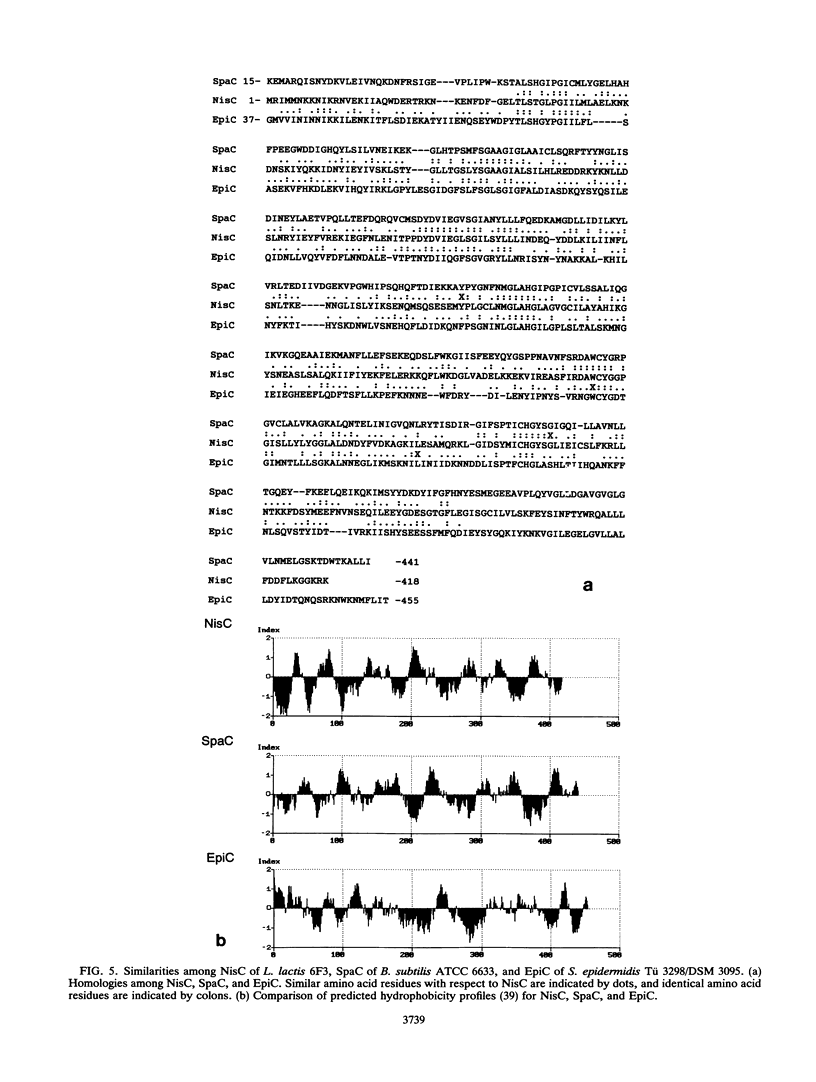
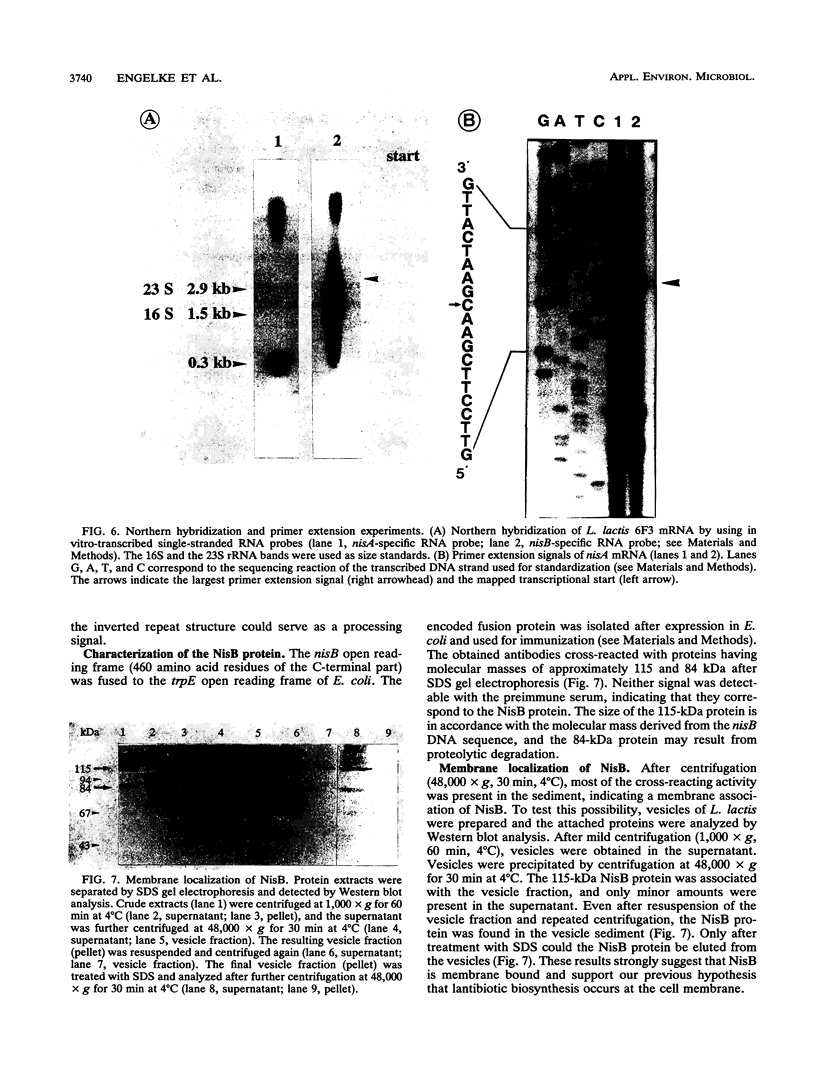
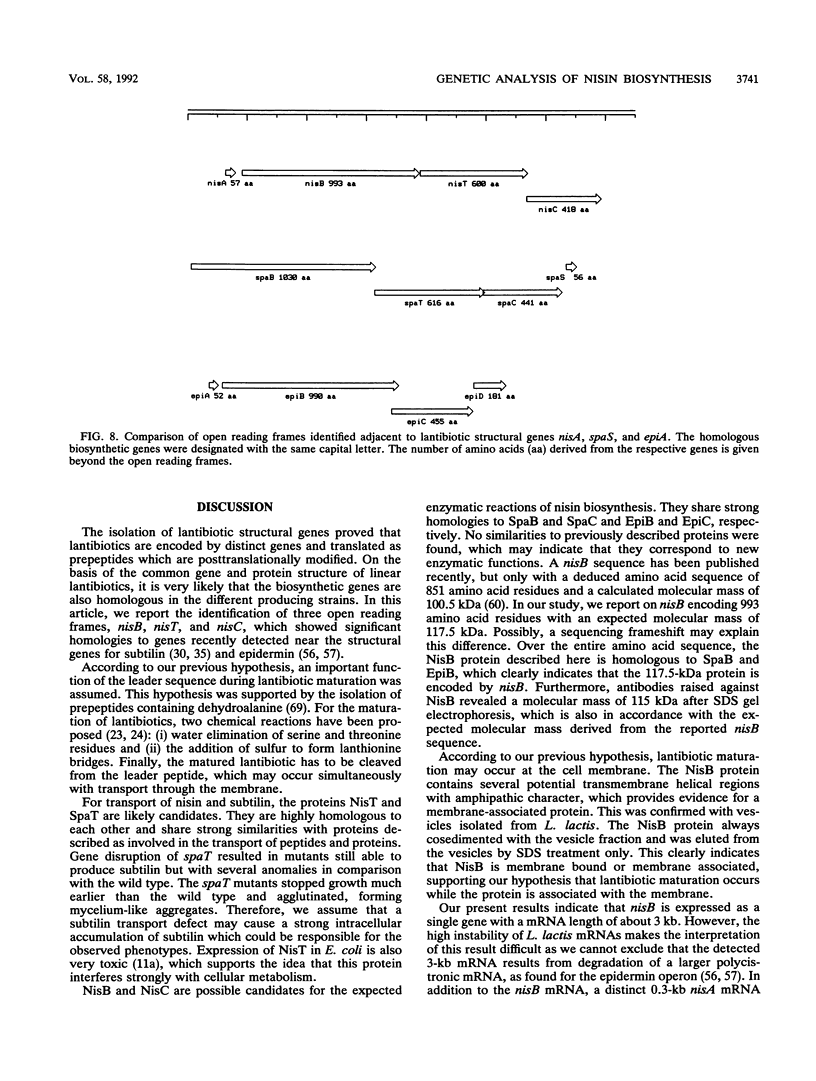
Nisin produced by Lactococcus lactis 6F3 is used as a food preservative and is the most important member of a group of peptide-antibiotics containing lanthionine bridges (lantibiotics) (N. Schnell, K.-D. Entian, U. Schneider, F. Götz, H. Zähner, R. Kellner, and G. Jung, Nature [London] 333:276-278, 1988). Nisin is ribosomally synthesized, and its structural gene, nisA, encodes a prepeptide that is posttranslationally modified, revealing the active lantibiotic (C. Kaletta and K.-D. Entian, J. Bacteriol. 171:1597-1601, 1989). Adjacent to nisA, the additional genes nisB, nisT, and nisC were identified. Over their entire sequences, these genes were homologous to genes recently identified as important for the biosynthesis of lantibiotics, that is, subtilin from Bacillus subtilis ATCC 6633 and epidermin from Staphylococcus epidermidis Tü 3298. Genes nisB, nisT, and nisC corresponded to open reading frames of 993, 600, and 418 amino acid residues, respectively. The nisT open reading frame is homologous to proteins of the HlyB (hemolysin B protein of Escherichia coli) subfamily. Proteins of this subfamily are responsible for the secretion of a variety of compounds, including large polypeptides, polysaccharides, and anti-drug tumors, indicating that NisT may be involved in nisin transport. Northern (RNA) blot analysis revealed a 0.3-kb transcript for the nisA structural gene, and the transcriptional start point of the nisA gene was determined by primer extension. Additionally, a mRNA of at least 3 kb was identified by using a hybridization probe specific to nisB. Antibodies were raised against the NisB protein, and Western blot (immunoblot) analysis revealed a molecular weight of about 115 kDa, which is in accordance with the theoretical protein size of 117.5 kDa as calculated from the nisB open reading frame. Several amphipathic transmembrane alpha-helices indicated that NisB is associated with the membrane. This was confirmed by preparing L. lactis vesicles. The NisB protein was tightly associated with the vesicle fraction and was released by sodium dodecyl sulfate treatment only. These results suggest that NisB is membrane associated and that nisin biosynthesis occurs at the cell membrane.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allgaier H., Jung G., Werner R. G., Schneider U., Zähner H. Epidermin: sequencing of a heterodetic tetracyclic 21-peptide amide antibiotic. Eur J Biochem. 1986 Oct 1;160(1):9–22. doi: 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- Augustin J., Rosenstein R., Wieland B., Schneider U., Schnell N., Engelke G., Entian K. D., Götz F. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992 Mar 15;204(3):1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Hansen J. N. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem. 1988 Jul 5;263(19):9508–9514. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight M. A., Holland I. B. Structure and function of haemolysin B,P-glycoprotein and other members of a novel family of membrane translocators. Mol Microbiol. 1990 Jun;4(6):873–880. doi: 10.1111/j.1365-2958.1990.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Buchman G. W., Banerjee S., Hansen J. N. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988 Nov 5;263(31):16260–16266. [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Welch R. A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985 Jul;163(1):94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P., Sakamoto H., Bellalou J., Ullmann A., Danchin A. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 1988 Dec 1;7(12):3997–4004. doi: 10.1002/j.1460-2075.1988.tb03288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros P., Croop J., Housman D. Mammalian multidrug resistance gene: complete cDNA sequence indicates strong homology to bacterial transport proteins. Cell. 1986 Nov 7;47(3):371–380. doi: 10.1016/0092-8674(86)90594-5. [DOI] [PubMed] [Google Scholar]
- Gross E., Kiltz H. H. The number and nature of , -unsaturated amino acids in subtilin. Biochem Biophys Res Commun. 1973 Jan 23;50(2):559–565. doi: 10.1016/0006-291x(73)90876-0. [DOI] [PubMed] [Google Scholar]
- Hancock K., Tsang V. C. India ink staining of proteins on nitrocellulose paper. Anal Biochem. 1983 Aug;133(1):157–162. doi: 10.1016/0003-2697(83)90237-3. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hui F. M., Morrison D. A. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J Bacteriol. 1991 Jan;173(1):372–381. doi: 10.1128/jb.173.1.372-381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram L. C. Synthesis of the antibiotic nisin: formation of lanthionine and beta-methyl-lanthionine. Biochim Biophys Acta. 1969 Jun 17;184(1):216–219. doi: 10.1016/0304-4165(69)90121-4. [DOI] [PubMed] [Google Scholar]
- Ingram L. A ribosomal mechanism for synthesis of peptides related to nisin. Biochim Biophys Acta. 1970 Nov 12;224(1):263–265. doi: 10.1016/0005-2787(70)90642-8. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D., Jung G. Prepeptide sequence of cinnamycin (Ro 09-0198): the first structural gene of a duramycin-type lantibiotic. Eur J Biochem. 1991 Jul 15;199(2):411–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D., Kellner R., Jung G., Reis M., Sahl H. G. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch Microbiol. 1989;152(1):16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- Kaletta C., Entian K. D. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol. 1989 Mar;171(3):1597–1601. doi: 10.1128/jb.171.3.1597-1601.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner R., Jung G., Hörner T., Zähner H., Schnell N., Entian K. D., Götz F. Gallidermin: a new lanthionine-containing polypeptide antibiotic. Eur J Biochem. 1988 Oct 15;177(1):53–59. doi: 10.1111/j.1432-1033.1988.tb14344.x. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Kaletta C., Schnell N., Entian K. D. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol. 1992 Jan;58(1):132–142. doi: 10.1128/aem.58.1.132-142.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Koivula T., Sibakov M., Palva I. Isolation and characterization of Lactococcus lactis subsp. lactis promoters. Appl Environ Microbiol. 1991 Feb;57(2):333–340. doi: 10.1128/aem.57.2.333-340.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse H., Hurst A. Preparation of spheroplasts from Streptococcus lactis. Can J Microbiol. 1972 Jun;18(6):825–831. doi: 10.1139/m72-128. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse N., Tenmyo O., Tomita K., Konishi M., Miyaki T., Kawaguchi H., Fukase K., Wakamiya T., Shiba T. Lanthiopeptin, a new peptide antibiotic. Production, isolation and properties of lanthiopeptin. J Antibiot (Tokyo) 1989 Jun;42(6):837–845. doi: 10.7164/antibiotics.42.837. [DOI] [PubMed] [Google Scholar]
- Otto R., Lageveen R. G., Veldkamp H., Konings W. N. Lactate efflux-induced electrical potential in membrane vesicles of Streptococcus cremoris. J Bacteriol. 1982 Feb;149(2):733–738. doi: 10.1128/jb.149.2.733-738.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger P., Seidel H., Tschesche H., Hammes W. P. The effect of nisin on murein synthesis. Arch Microbiol. 1980 Oct;127(3):187–193. doi: 10.1007/BF00427192. [DOI] [PubMed] [Google Scholar]
- Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989 Sep 8;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Rogers L. A. THE INHIBITING EFFECT OF STREPTOCOCCUS LACTIS ON LACTOBACILLUS BULGARICUS. J Bacteriol. 1928 Nov;16(5):321–325. doi: 10.1128/jb.16.5.321-325.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl H. G., Brandis H. Production, purification and chemical properties of an antistaphylococcal agent produced by Staphylococcus epidermidis. J Gen Microbiol. 1981 Dec;127(2):377–384. doi: 10.1099/00221287-127-2-377. [DOI] [PubMed] [Google Scholar]
- Sahl H. G. Influence of the staphylococcinlike peptide Pep 5 on membrane potential of bacterial cells and cytoplasmic membrane vesicles. J Bacteriol. 1985 May;162(2):833–836. doi: 10.1128/jb.162.2.833-836.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl H. G., Kordel M., Benz R. Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch Microbiol. 1987;149(2):120–124. doi: 10.1007/BF00425076. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell N., Engelke G., Augustin J., Rosenstein R., Ungermann V., Götz F., Entian K. D. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem. 1992 Feb 15;204(1):57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K. D., Götz F., Hörner T., Kellner R., Jung G. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol Lett. 1989 Apr;49(2-3):263–267. doi: 10.1016/0378-1097(89)90050-5. [DOI] [PubMed] [Google Scholar]
- Schnell N., Entian K. D., Schneider U., Götz F., Zähner H., Kellner R., Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988 May 19;333(6170):276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- Steen M. T., Chung Y. J., Hansen J. N. Characterization of the nisin gene as part of a polycistronic operon in the chromosome of Lactococcus lactis ATCC 11454. Appl Environ Microbiol. 1991 Apr;57(4):1181–1188. doi: 10.1128/aem.57.4.1181-1188.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee C. A., Lo R. Y. Cloning, nucleotide sequence, and characterization of genes encoding the secretion function of the Pasteurella haemolytica leukotoxin determinant. J Bacteriol. 1989 Feb;171(2):916–928. doi: 10.1128/jb.171.2.916-928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Lundström K., Lehtovaara P., Sarvas M., Ruohonen M., Palva I. Transcription and translation of foreign genes in Bacillus subtilis by the aid of a secretion vector. J Bacteriol. 1985 Apr;162(1):176–182. doi: 10.1128/jb.162.1.176-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Ven F. J., Van den Hooven H. W., Konings R. N., Hilbers C. W. NMR studies of lantibiotics. The structure of nisin in aqueous solution. Eur J Biochem. 1991 Dec 18;202(3):1181–1188. doi: 10.1111/j.1432-1033.1991.tb16488.x. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil H. P., Beck-Sickinger A. G., Metzger J., Stevanovic S., Jung G., Josten M., Sahl H. G. Biosynthesis of the lantibiotic Pep5. Isolation and characterization of a prepeptide containing dehydroamino acids. Eur J Biochem. 1990 Nov 26;194(1):217–223. doi: 10.1111/j.1432-1033.1990.tb19446.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]