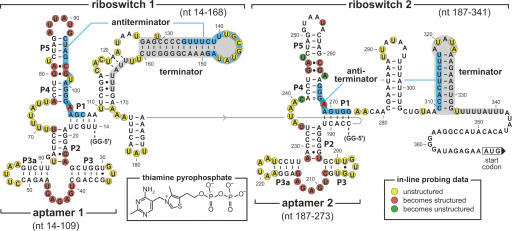
FIGURE 3.
Sequence and secondary structure architecture of the tandem TPP riboswitches from the 5′ UTR of the tenA operon of B. anthracis. Aptamer models are based on phylogenetic (Mironov et al. 2002; Winkler et al. 2002b) and X-ray structure (Serganov et al. 2006; Thore et al. 2006) data. Ranges of nucleotides identify the various constructs used for biochemical analyses. Putative antiterminator structures (blue-shaded nucleotides), putative terminator structures (gray-shaded nucleotides), two G residues (GG-5′) were added to the 5′ end of constructs to improve yields from in vitro transcription. In-line probing markings reflect data generated (not shown) using constructs Thi1+T1 (riboswitch 1) and Thi2+T2 (riboswitch 2).