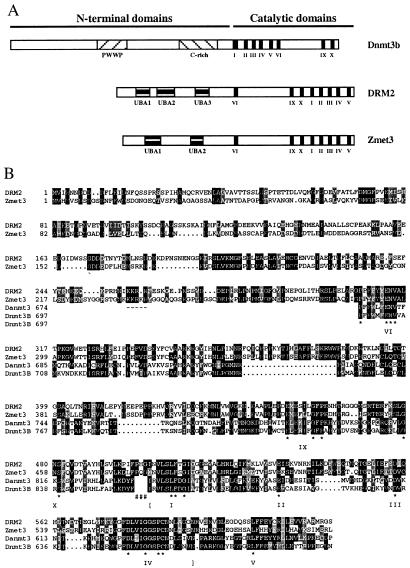
Figure 1.
(A) Schematic diagram of the domain structures of Dnmt3b, DRM2, and Zmet3. Figure is drawn to scale. Shaded boxes show the different motifs present in these proteins, including the PWWP and cysteine rich (C-rich) motifs present in Dnmt3b and the UBA domains present in DRM2 and Zmet3. Roman numerals denote the motifs of the methyltransferase catalytic domains. (B) Alignment of DRM2 of Arabidopsis thaliana with Zmet3 of Zea mays and the methyltransferase catalytic domains of mouse Dnmt3b (GenBank accession no. AF068628) and Danio rerio Zmt3 (Danmt3; accession no. AF135438). Alignments were done in clustalX 1.8 using default parameters and shaded in MacBoxShade 1.0.8 (Michael D. Baron, Institute for Animal Health, Surrey, U.K.). Identical residues are shown with a black background and similar residues with a gray background. Dashes show putative nuclear localization sequences that are conserved in the plant proteins. Pound symbols (#) show the point of rearrangement of the plant proteins relative to the animal proteins. Here the numbering of the animal methyltransferases begins at amino acid 581 for Dnmt3b and 558 for Danmt3. Conserved catalytic motifs I–VI and IX–X are marked. Asterisks denote conserved amino acids present in each motif (11). Brackets show the exact region used in the alignments that were used to produce the tree shown in Fig. 3.