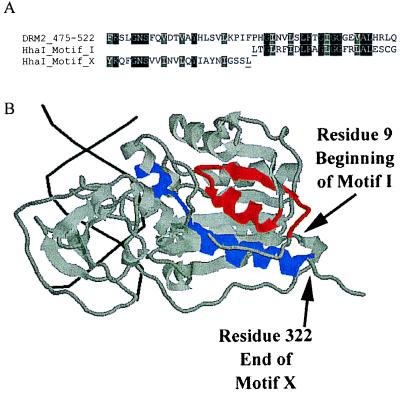
Figure 2.
(A) An alignment of motifs X and I of HhaI with the sequence of DRM2 spanning the point of motif rearrangement. HhaI residues underlined are the last residue of motif X and the first residue of motif I and correspond the positions in B marked by arrows. Alignments were done in clustalX and shaded in boxshade. (B) rasmol-generated image (Roger Sayle, Glaxo Research and Development, Greenford, Middlesex, U.K.) of Protein Database ID 5MHT (ternary structure of HhaI methyltransferase with hemimethylated DNA and S-adenosylhomocysteine) (36, 37). DNA helix is shown in black, the majority of HhaI is shaded gray, motif I is red, and motif X is blue. The colored regions correspond to those shown in the alignment in A, which are residues 9–32 for motif I and residues 299–322 for motif X. Arrows shows the last residue of motif X and first residue of motif I.