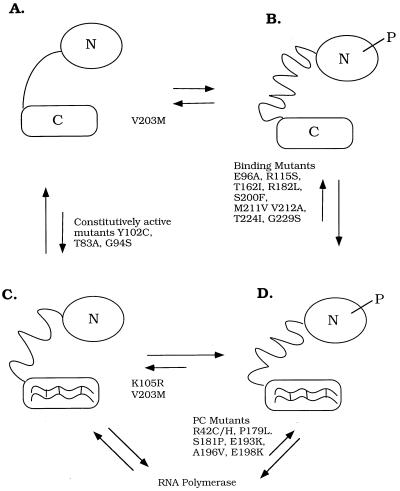
Figure 6.
Four-states model for OmpR. (A and C) The unphosphorylated forms of OmpR. (B and D) The phosphorylated forms of OmpR. (A and B) The forms of OmpR that are not bound to DNA. (C and D) The DNA-bound forms of OmpR. Relevant mutants are shown next to the arrows denoting the equilibria that seem to be affected by the substitution. The OmpR mutant K105R (C) binds to DNA with an affinity similar to unphosphorylated OmpR, but the mutant is not stably phosphorylated (15). DNA-binding mutants shown are E96A and R115S (48). They can be phosphorylated by EnvZ-P; however, they do not bind ompF and ompC, and they have an F−C− phenotype. The DNA binding mutants T162I, R182L, S200F, T224I, and G229S (49) and the double-mutant M211V/V212A (50) are defective in binding. Their ability to be phosphorylated has not been determined but is presumed to be unaffected by the mutation or mutations. The V203M mutant is affected in binding at ompC and at some ompF sites, and its phenotype is FcC− (51–53). Surprisingly, phosphorylation by acetyl phosphate of this mutant is inhibited (V. Tran and L.J.K., unpublished results), and it is therefore shown locked in the A/C position. The constitutively active mutants Y102C (54), T83A, and G94S (55) bind DNA with high affinity in the absence of phosphorylation. The activation mutants S181P (49), P179L, A196V, E198K, R42C/H (56), and E193K (52) bind but cannot activate transcription and are presumed to be defective in their ability to interact with RNA polymerase. Note that the linker is represented as being in a different conformation in each of the four states. N = N terminus; C = C terminus, P = phosphoryl group.