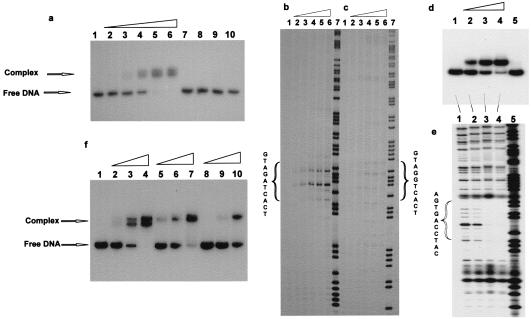
Figure 2.
Binding of pseudocomplementary PNAs to double-stranded DNA. (a) PNAs 1495 and 1496 (see Table 1) were incubated for 16 hr at 37°C in 10 mM sodium phosphate/1 mM EDTA buffer, pH 7.0, with a 204-bp DNA fragment (HindIII/PvuII fragment of p206 3′-32P-end-labeled at the HindIII site) containing the 10-bp PNA target. The following concentrations were used: lanes 1–6, 0, 5, 10, 20, 40, and 80 nM each of PNAs 1495 and 1496; lanes 7 and 9, 40 and 80 nM, respectively, of PNA 1495; and lanes 8 and 10, 40 and 80 nM, respectively, of PNA 1496. The presence of two shifted complexes is not understood. HPLC and MALDI-TOF analyses of the PNAs did not indicate heterogeneity, but it could be that there are two slowly exchanging “structural isomers” of this very “crowded” double duplex invasion complex. (b and c) Sequence specificity of the PNA binding. PNAs 1495 and 1496 were bound to the HindIII/PvuII fragment of p206 (b) or p259 (one mismatch target) (c) as described above. Subsequently the complexes were probed with KMnO4 and the samples were analyzed by electrophoresis in polyacrylamide sequencing gels followed by autoradiography. The PNA concentrations in lanes 1–7 were 0, 60, 200, and 600 nM and 2 mM and 6 mM. Lane 7 is an A/G sequence marker. The 10-mer PNA targets are indicated. (d) Experiment as in a except that the PNA pairs 1837/1838 (H-Lys-GsUDGGsUCDCsU-Lys-NH2/H-Lys-DGsUGDCCsUDC-Lys-NH2) and the plasmid p259 were used. The PNA concentrations were as follows: lane 1, control without PNA; lanes 2–5, 30 nM PNA 1838 and 100 nM, 150 nM, 200 nM, and 0 nM of PNA 1837, respectively. (e) DNase I probing (after adjusting the buffer to 50 mM Tris⋅HCl, pH 7.5/1 mM Mg2Cl) of samples 1–4 as described for d; lane 5 is an A/G sequence marker. (f) Experiment as in a except that the PNA pairs 1495/1496 (60% AT modification) (lanes 2–4), 1674/1672 (H-Lys-GsUDGDsUCDCT-Lys-NH2/H-Lys-AGsUGDsUCsUDC-Lys-NH2) (50% AT modification) (lanes 5–7), and 1675/1673 (H-Lys-GsUAGDsUCDCT-Lys-NH2/H-Lys-AGsUGDsUCTDC-Lys-NH2) (40% AT modification) (lanes 8–10) were used at concentrations of 5 nM (lane 2), 15 nM (lanes 3, 5, and 8), 45 nM (lanes 4, 6, and 9), or 150 nM (lanes 7 and 10). Lane 1 is a control without PNA.