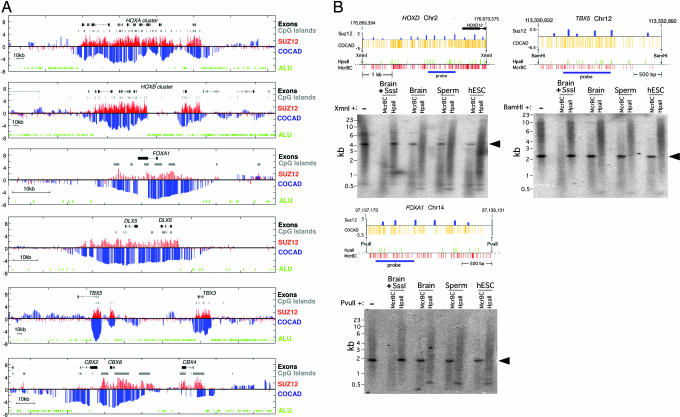
Fig. 4.
Hyperconserved CpG domains correspond to unmethylated PRC2-binding domains. (A) Genomic regions around several key developmental genes (drawn to scale; exons are marked as thick lines). For each genomic region, we plot the Suz12 raw ChIP-binding ratio (red) alongside the COCAD CpG conservation score (blue). Also plotted are Alu elements (green) and CpG islands (gray). The COCAD and Suz12 profiles, although obtained by unrelated methods, show strong similarity. The observed correlation is particularly significant because for the large majority of the genome, both the COCAD scores and Suz12 ChIP intensities are not significantly different from 0 (see http://www.pnas.org/cgi/content/full/0609746104/DC1SI Text). (B) HCGDs are unmethylated in human ES cells, sperm, and brain. Resistance to McrBC (a methylation-dependent endonuclease complex) and sensitivity to HpaII (a methylation-inhibited restriction endonuclease) show that regions with low COCAD scores and high PRC2 association are unmethylated at all stages tested. Distributions of McrBC and HpaII recognition sequences are shown. Prior methylation of DNA at all CpG dinucleotides with SssI DNA methyltransferase renders DNA resistant to HpaII and sensitive to McrBC, whereas DNA from human ES cells, sperm, and brain is sensitive to HpaII and resistant to McrBC and therefore unmethylated at all or nearly all CpG dinucleotides at the regions indicated. The hybridization background is an artifact of the very high G + C contents of the probe and target sequences. Arrowheads indicate unmethylated domains in DNA from brain, sperm, and human ES cells.