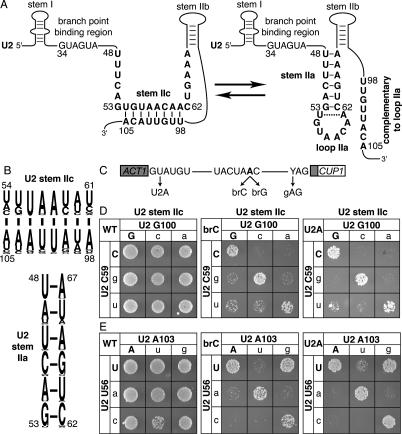
Figure 1.
Disrupting U2 stem IIc exacerbates the splicing defect of a substrate mutated at the branch site or the 5′ splice site. (A) Structure of U2 stem IIc and stem–loop IIa. Nucleotides involved in stem IIc, stem IIa, and loop IIa are bold. The nucleotides of U2 that bind the intronic branch site consensus sequence are shown. Nucleotide sequence and numbers correspond to the Saccharomyces cerevisiae sequence. The dashed line marks a nonconserved base pair that can extend stem IIa in S. cerevisiae. (B) A pictogram of stem IIc and IIa showing the consensus sequence for organisms ranging from budding yeast to humans; numbering refers to the budding yeast sequence. The height of the letter is proportional to the frequency of the nucleotide in the alignment. (C) Schematic diagram of the ACT1-CUP1 splicing reporter, which when spliced confers copper resistance to budding yeast (Lesser and Guthrie 1993). The intron consensus sequences for budding yeast are shown along with point mutations used in this study. The branch site adenosine is bold. (D,E) Mutations that disrupt base-pairing in U2 stem IIc enhance the growth defect conferred by the 5′ splice site mutation U2A and the branch site mutation brC. Compensatory analysis by copper resistance of U2 stem IIc base pairs C59/G100 (D) and U56/A103 (E) is shown. The matrices show the copper resistance of the wild-type (WT; left), brC (middle), or U2A (right) ACT1-CUP1 splicing reporters in yJPS1035 expressing U2 variants having single or double mutations in the stem IIc base pairs. Cells were grown for 3–4 d at 30°C on solid media containing 0.05 mM copper sulfate. Wild-type residues are capitalized.