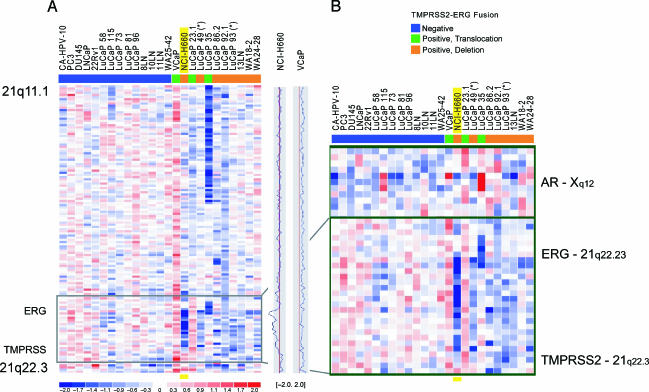
Figure 3.
Genomic loss between TMPRSS2 and ERG on chromosome 21q22.2-3 in prostate cancer cell lines, xenografts, and metastatic prostate cancer samples. 8K SNP data on a panel of 25 prostate cancer samples revealed genomic deletion between TMPRSS2 and ERG (21q22.2-3) in a subset of samples. (A) Twenty-five prostate cancer samples, including 7 cell lines, 11 xenografts, and 7 prostate cancer metastases, were analyzed for their TMPRSS2-ERG fusion status by qPCR and/or FISH and color-coded as described before [3] (blue, fusion-negative; green, fusion-positive through translocation; orange, fusion-positive through deletion). The plots on the right side of the panel represent the copy number ratio of the NCI-H660 and VCaP cell lines (vertical red lines represent baseline; no copy number variation). It is evident that VCaP shows copy number gain throughout the whole q arm of chromosome 21. (B) Magnification of the black framed box in (A), and status of the AR on chromosome X in these 25 samples. The blue signal in NCI-H660 corresponds to genomic copy number loss between TMPRSS2 and ERG. The strong intensity of this signal is consistent with homozygous loss, as demonstrated by FISH. The boundaries of the intronic deletion of NCI-H660 are very similar to the deletions seen in xenografts and tissue samples. Toward the telomeric side of the deletion in NCI-H660, the signal intensity is weaker, confirming different lengths of the deletions seen by FISH. The androgen-independent NCI-H660 cell line shows a loss in the region of the AR, whereas VCaP, which is known to be androgen-responsive, shows genomic gain in this area.