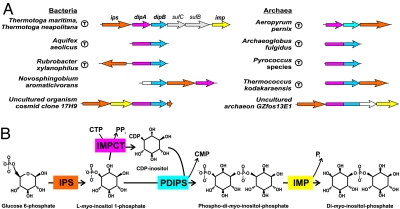
Fig. 1.
Reconstruction of the DIP biosynthesis pathway in prokaryotic genomes. (A) Genome context of DIP pathway genes in bacterial and archaeal genomes. (Hyper)thermophilic species are marked by a circle with “T.” Orthologs are shown by matching colors of individual genes or respective segments of fused genes. Predicted novel genes (dipA and dipB) are fused (dipAB) or occur in chromosomal clusters with each other and, often, with the two known genes of the DIP pathway (ips and imp). (B) A diagram of the revised DIP synthesis pathway. IPS, EC 5.5.1.4; IMPCT, 2.7.7.–; PDIPS, 2.7.8.–; IMP, 3.1.3.25.