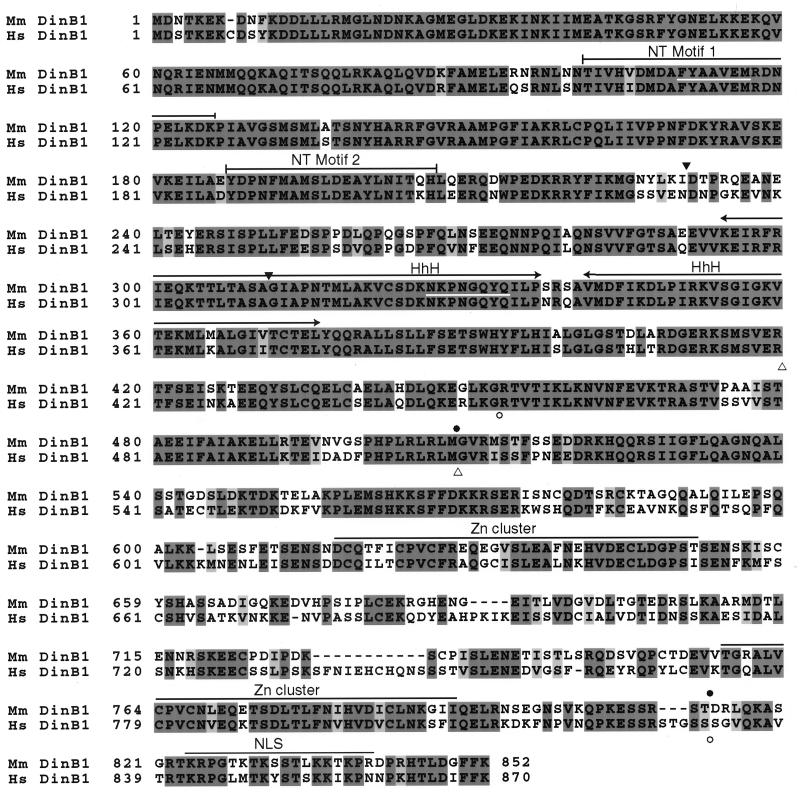
Figure 1.
Amino acid alignment of the mouse (Mus musculus; Mm) and human (Homo sapiens; Hs) DinB proteins. The alignment was generated with clustalw. Identical residues are shaded in dark gray. Similar residues are shaded in light gray. Sites in the mouse DinB1 protein sequence corresponding to the degenerate primers used for cloning are underlined in white (lines 2 and 6 of the paired sequences). Putative functional units are overlined: NT, nucleotidyl transferase; HhH, helix–hairpin–helix; Zn cluster, modified Zn cluster; NLS, bipartite nuclear localization signal. The positions of the predicted deletions in the two isoforms of the mouse [amino acid positions 231–310 (▾) and 509–813 (●)] and human [amino acid positions 420–509 (▵) and 453–831 (○)] DinB1 proteins resulting from alternative splicing of transcripts in testis are indicated.