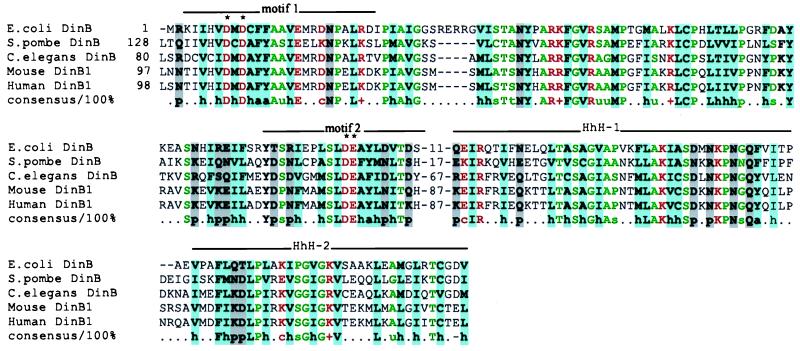
Figure 2.
Conserved motifs within the DinB branch of the UmuC/DinB superfamily. The two conserved motifs containing the putative catalytic residues mentioned in the text (marked by asterisks) and the two HhH modules are overlined. Gene identification numbers are, for E. coli, 2501652; for Schizosaccharomyces pombe, 4038629; and, for C. elegans, 465873. The consensus that includes residues conserved in all aligned sequences is shown beneath the alignment with the following designations: a, aromatic (F, Y, and W); h, hydrophobic (A, C, I, L, V, M, F, Y, and W); p, polar (D, E, N, Q, K, R, H, S, and T); c, charged (D, E, K, and R); +, positively charged (K and R); −, negatively charged (D and E); s, small (G, A, S, P, V, D, and N); and u, tiny (G, A, and S). Conserved residues are color coded according to the consensus.